

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Tyrosine-protein kinase ITK/TSK | ||
| Ligand | BDBM50037072 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1435180 (CHEMBL3389531) | ||
| Ki | 20±n/a nM | ||
| Citation |  Trani, G; Barker, JJ; Bromidge, SM; Brookfield, FA; Burch, JD; Chen, Y; Eigenbrot, C; Heifetz, A; Ismaili, MH; Johnson, A; Krülle, TM; MacKinnon, CH; Maghames, R; McEwan, PA; Montalbetti, CA; Ortwine, DF; Pérez-Fuertes, Y; Vaidya, DG; Wang, X; Zarrin, AA; Pei, Z Design, synthesis and structure-activity relationships of a novel class of sulfonylpyridine inhibitors of Interleukin-2 inducible T-cell kinase (ITK). Bioorg Med Chem Lett24:5818-23 (2014) [PubMed] Article Trani, G; Barker, JJ; Bromidge, SM; Brookfield, FA; Burch, JD; Chen, Y; Eigenbrot, C; Heifetz, A; Ismaili, MH; Johnson, A; Krülle, TM; MacKinnon, CH; Maghames, R; McEwan, PA; Montalbetti, CA; Ortwine, DF; Pérez-Fuertes, Y; Vaidya, DG; Wang, X; Zarrin, AA; Pei, Z Design, synthesis and structure-activity relationships of a novel class of sulfonylpyridine inhibitors of Interleukin-2 inducible T-cell kinase (ITK). Bioorg Med Chem Lett24:5818-23 (2014) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Tyrosine-protein kinase ITK/TSK | |||
| Name: | Tyrosine-protein kinase ITK/TSK | ||
| Synonyms: | EMT | ITK | ITK_HUMAN | Kinase EMT | LYK | T-cell-specific kinase | Tyrosine-protein kinase ITK | Tyrosine-protein kinase ITK (ITK) | Tyrosine-protein kinase Lyk | ||
| Type: | Protein | ||
| Mol. Mass.: | 71839.20 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q08881 | ||
| Residue: | 620 | ||
| Sequence: |
| ||
| BDBM50037072 | |||
| n/a | |||
| Name | BDBM50037072 | ||
| Synonyms: | CHEMBL3355734 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C27H26F3N5O3S | ||
| Mol. Mass. | 557.587 | ||
| SMILES | O[C@H]1CC[C@@H](CC1)Nc1cc(cc(Nc2cc([nH]n2)-c2ccc(cc2)C(F)(F)F)n1)S(=O)(=O)c1ccccc1 |r,wU:4.7,wD:1.0,(7.54,-17.72,;7.54,-16.18,;8.88,-15.41,;8.88,-13.86,;7.55,-13.11,;6.21,-13.87,;6.21,-15.4,;7.55,-11.57,;6.21,-10.8,;4.88,-11.57,;3.54,-10.8,;3.54,-9.26,;4.87,-8.48,;4.87,-6.94,;6.2,-6.17,;7.61,-6.8,;8.64,-5.65,;7.87,-4.32,;6.36,-4.65,;10.17,-5.81,;10.8,-7.22,;12.33,-7.38,;13.23,-6.13,;12.6,-4.72,;11.07,-4.57,;14.77,-6.28,;15.4,-7.68,;15.67,-5.03,;16.3,-6.28,;6.21,-9.25,;2.2,-11.57,;2.96,-12.9,;1.43,-12.89,;.87,-10.8,;.88,-9.26,;-.45,-8.49,;-1.79,-9.26,;-1.78,-10.8,;-.46,-11.57,)| | ||
| Structure | 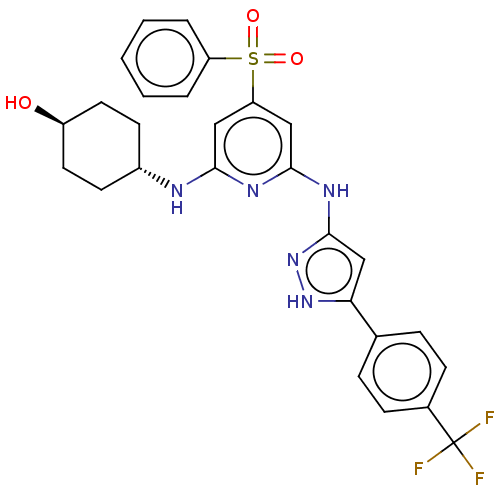
| ||