

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Macrophage-stimulating protein receptor | ||
| Ligand | BDBM50103899 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1505178 (CHEMBL3594720) | ||
| IC50 | 270±n/a nM | ||
| Citation |  Liao, W; Hu, G; Guo, Z; Sun, D; Zhang, L; Bu, Y; Li, Y; Liu, Y; Gong, P Design and biological evaluation of novel 4-(2-fluorophenoxy)quinoline derivatives bearing an imidazolone moiety as c-Met kinase inhibitors. Bioorg Med Chem23:4410-22 (2015) [PubMed] Article Liao, W; Hu, G; Guo, Z; Sun, D; Zhang, L; Bu, Y; Li, Y; Liu, Y; Gong, P Design and biological evaluation of novel 4-(2-fluorophenoxy)quinoline derivatives bearing an imidazolone moiety as c-Met kinase inhibitors. Bioorg Med Chem23:4410-22 (2015) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Macrophage-stimulating protein receptor | |||
| Name: | Macrophage-stimulating protein receptor | ||
| Synonyms: | 2.7.10.1 | CD_antigen=CD136 | CDw136 | MSP receptor | MST1R | Macrophage-stimulating protein receptor (MST1R) | Macrophage-stimulating protein receptor alpha chain | Macrophage-stimulating protein receptor beta chain | PTK8 | Protein-tyrosine kinase 8 | RON | RON_HUMAN | Tyrosine kinase receptor ron | p185-Ron | ||
| Type: | Protein | ||
| Mol. Mass.: | 152270.76 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q04912 | ||
| Residue: | 1400 | ||
| Sequence: |
| ||
| BDBM50103899 | |||
| n/a | |||
| Name | BDBM50103899 | ||
| Synonyms: | CHEMBL3594105 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C35H36ClFN6O5 | ||
| Mol. Mass. | 675.149 | ||
| SMILES | COc1cc2c(Oc3ccc(NC(=O)c4[nH]c(=O)n(c4C)-c4ccccc4Cl)cc3F)ccnc2cc1OCCCN1CCN(C)CC1 |(3.99,-2.77,;3.99,-1.54,;2.66,-.77,;1.33,-1.54,;,-.77,;-1.33,-1.54,;-1.33,-3.08,;-2.66,-3.85,;-3.99,-3.09,;-5.32,-3.86,;-5.32,-5.4,;-6.65,-6.18,;-6.64,-7.72,;-5.58,-8.33,;-7.98,-8.49,;-9.36,-7.86,;-10.39,-9.01,;-11.62,-8.88,;-9.62,-10.34,;-8.11,-10.02,;-7.19,-10.83,;-10.24,-11.75,;-9.24,-12.92,;-9.75,-14.37,;-11.27,-14.65,;-12.27,-13.48,;-11.75,-12.03,;-12.55,-11.09,;-3.98,-6.17,;-2.65,-5.39,;-1.58,-6.01,;-2.68,-.77,;-2.68,.77,;-1.33,1.54,;,.77,;1.33,1.54,;2.66,.77,;3.99,1.54,;3.99,3.08,;5.33,3.86,;5.32,5.4,;6.66,6.17,;6.66,7.71,;7.99,8.48,;9.32,7.71,;10.39,8.33,;9.32,6.17,;7.99,5.4,)| | ||
| Structure | 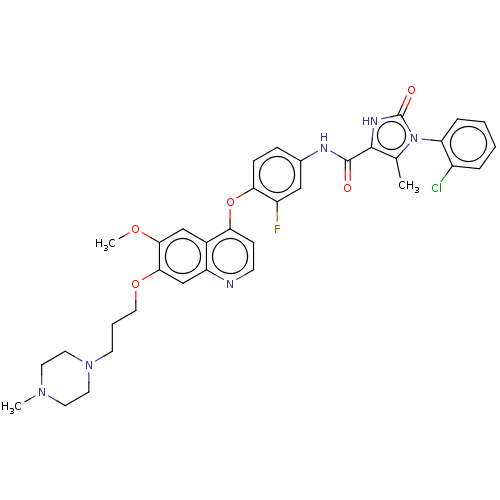
| ||