| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Glucagon-like peptide 1 receptor |
|---|
| Ligand | BDBM50130490 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1525355 (CHEMBL3636172) |
|---|
| EC50 | 470±n/a nM |
|---|
| Citation |  Swedberg, JE; Schroeder, CI; Mitchell, JM; Durek, T; Fairlie, DP; Edmonds, DJ; Griffith, DA; Ruggeri, RB; Derksen, DR; Loria, PM; Liras, S; Price, DA; Craik, DJ Cyclic alpha-conotoxin peptidomimetic chimeras as potent GLP-1R agonists. Eur J Med Chem103:175-84 (2015) [PubMed] Article Swedberg, JE; Schroeder, CI; Mitchell, JM; Durek, T; Fairlie, DP; Edmonds, DJ; Griffith, DA; Ruggeri, RB; Derksen, DR; Loria, PM; Liras, S; Price, DA; Craik, DJ Cyclic alpha-conotoxin peptidomimetic chimeras as potent GLP-1R agonists. Eur J Med Chem103:175-84 (2015) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Glucagon-like peptide 1 receptor |
|---|
| Name: | Glucagon-like peptide 1 receptor |
|---|
| Synonyms: | GLP-1 receptor | GLP-1-R | GLP-1R | GLP1R | GLP1R_HUMAN | Glucagon-like peptide 1 receptor | Glucagon-like peptide 1 receptor (GLP-1) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 53035.78 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P43220 |
|---|
| Residue: | 463 |
|---|
| Sequence: | MAGAPGPLRLALLLLGMVGRAGPRPQGATVSLWETVQKWREYRRQCQRSLTEDPPPATDL
FCNRTFDEYACWPDGEPGSFVNVSCPWYLPWASSVPQGHVYRFCTAEGLWLQKDNSSLPW
RDLSECEESKRGERSSPEEQLLFLYIIYTVGYALSFSALVIASAILLGFRHLHCTRNYIH
LNLFASFILRALSVFIKDAALKWMYSTAAQQHQWDGLLSYQDSLSCRLVFLLMQYCVAAN
YYWLLVEGVYLYTLLAFSVLSEQWIFRLYVSIGWGVPLLFVVPWGIVKYLYEDEGCWTRN
SNMNYWLIIRLPILFAIGVNFLIFVRVICIVVSKLKANLMCKTDIKCRLAKSTLTLIPLL
GTHEVIFAFVMDEHARGTLRFIKLFTELSFTSFQGLMVAILYCFVNNEVQLEFRKSWERW
RLEHLHIQRDSSMKPLKCPTSSLSSGATAGSSMYTATCQASCS
|
|
|
|---|
| BDBM50130490 |
|---|
| n/a |
|---|
| Name | BDBM50130490 |
|---|
| Synonyms: | CHEMBL3633851 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C113H142F2N24O31S2 |
|---|
| Mol. Mass. | 2434.606 |
|---|
| SMILES | [H][C@@]12CCCN1C(=O)[C@H](Cc1cnc[nH]1)NC(=O)[C@H](CO)NC(=O)[C@]1([H])CSSC[C@]([H])(NC(=O)[C@H](CCCc3ccccc3)NC(=O)[C@H](Cc3ccc(cc3)-c3ccc(OC)cc3CC)NC(=O)[C@H](CC(O)=O)NC(=O)[C@H](CO)NC(=O)[C@@]([H])(NC(=O)[C@](C)(Cc3c(F)cccc3F)NC(=O)[C@@]([H])(NC(=O)CNC(=O)[C@H](CCC(O)=O)NC2=O)[C@@H](C)O)[C@@H](C)O)C(=O)NCC(=O)N[C@@H](Cc2ccc(cc2)-c2ccccc2)C(=O)N[C@@H](C)C(=O)N[C@@H](C)C(=O)NCC(=O)NCC(=O)NCC(=O)N[C@@H](C)C(=O)N1 |r| |
|---|
| Structure | 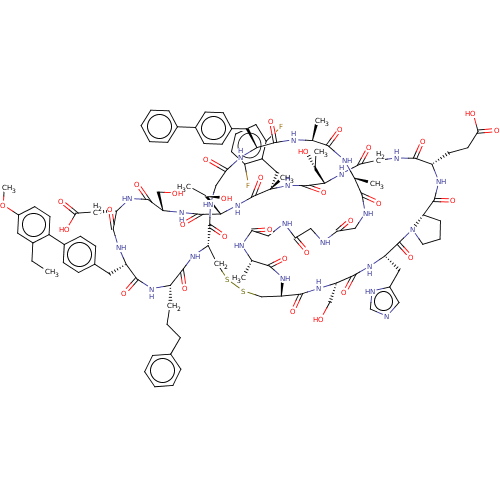
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Swedberg, JE; Schroeder, CI; Mitchell, JM; Durek, T; Fairlie, DP; Edmonds, DJ; Griffith, DA; Ruggeri, RB; Derksen, DR; Loria, PM; Liras, S; Price, DA; Craik, DJ Cyclic alpha-conotoxin peptidomimetic chimeras as potent GLP-1R agonists. Eur J Med Chem103:175-84 (2015) [PubMed] Article
Swedberg, JE; Schroeder, CI; Mitchell, JM; Durek, T; Fairlie, DP; Edmonds, DJ; Griffith, DA; Ruggeri, RB; Derksen, DR; Loria, PM; Liras, S; Price, DA; Craik, DJ Cyclic alpha-conotoxin peptidomimetic chimeras as potent GLP-1R agonists. Eur J Med Chem103:175-84 (2015) [PubMed] Article