| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Potassium voltage-gated channel subfamily H member 2 |
|---|
| Ligand | BDBM50028854 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1552430 (CHEMBL3761681) |
|---|
| Ki | 41300±n/a nM |
|---|
| Citation |  Schiffler, MA; Antonysamy, S; Bhattachar, SN; Campanale, KM; Chandrasekhar, S; Condon, B; Desai, PV; Fisher, MJ; Groshong, C; Harvey, A; Hickey, MJ; Hughes, NE; Jones, SA; Kim, EJ; Kuklish, SL; Luz, JG; Norman, BH; Rathmell, RE; Rizzo, JR; Seng, TW; Thibodeaux, SJ; Woods, TA; York, JS; Yu, XP Discovery and Characterization of 2-Acylaminoimidazole Microsomal Prostaglandin E Synthase-1 Inhibitors. J Med Chem59:194-205 (2016) [PubMed] Article Schiffler, MA; Antonysamy, S; Bhattachar, SN; Campanale, KM; Chandrasekhar, S; Condon, B; Desai, PV; Fisher, MJ; Groshong, C; Harvey, A; Hickey, MJ; Hughes, NE; Jones, SA; Kim, EJ; Kuklish, SL; Luz, JG; Norman, BH; Rathmell, RE; Rizzo, JR; Seng, TW; Thibodeaux, SJ; Woods, TA; York, JS; Yu, XP Discovery and Characterization of 2-Acylaminoimidazole Microsomal Prostaglandin E Synthase-1 Inhibitors. J Med Chem59:194-205 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Potassium voltage-gated channel subfamily H member 2 |
|---|
| Name: | Potassium voltage-gated channel subfamily H member 2 |
|---|
| Synonyms: | 1,3-beta-glucan synthase component GLS2 | Cytochrome P450 3A4 | ERG | ERG1 | Eag-related protein 1 | Ether a-go-go related gene potassium channel (hERG) | Ether-a-go-go-related gene (HERG) | Ether-a-go-go-related gene potassium channel (hERG) | Ether-a-go-go-related gene potassium channel 1 | Ether-a-go-go-related gene potassium channel 1 (HERG) | Ether-a-go-go-related gene potassium channel 1 (hERG1) | Ether-a-go-go-related protein (hERG) | Ether-a-go-go-related protein 1 | Ether-a-go-go-related protein 1 (HERG) | H-ERG | HERG | KCNH2 | KCNH2_HUMAN | Potassium voltage-gated channel subfamily H member 2 (hERG) | Transcriptional regulator ERG | Voltage-gated potassium channel subunit Kv11.1 | eag homolog | hERG Potassium Channel 1 | putative potassium channel subunit |
|---|
| Type: | Multi-pass membrane protein |
|---|
| Mol. Mass.: | 126672.65 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q12809 |
|---|
| Residue: | 1159 |
|---|
| Sequence: | MPVRRGHVAPQNTFLDTIIRKFEGQSRKFIIANARVENCAVIYCNDGFCELCGYSRAEVM
QRPCTCDFLHGPRTQRRAAAQIAQALLGAEERKVEIAFYRKDGSCFLCLVDVVPVKNEDG
AVIMFILNFEVVMEKDMVGSPAHDTNHRGPPTSWLAPGRAKTFRLKLPALLALTARESSV
RSGGAGGAGAPGAVVVDVDLTPAAPSSESLALDEVTAMDNHVAGLGPAEERRALVGPGSP
PRSAPGQLPSPRAHSLNPDASGSSCSLARTRSRESCASVRRASSADDIEAMRAGVLPPPP
RHASTGAMHPLRSGLLNSTSDSDLVRYRTISKIPQITLNFVDLKGDPFLASPTSDREIIA
PKIKERTHNVTEKVTQVLSLGADVLPEYKLQAPRIHRWTILHYSPFKAVWDWLILLLVIY
TAVFTPYSAAFLLKETEEGPPATECGYACQPLAVVDLIVDIMFIVDILINFRTTYVNANE
EVVSHPGRIAVHYFKGWFLIDMVAAIPFDLLIFGSGSEELIGLLKTARLLRLVRVARKLD
RYSEYGAAVLFLLMCTFALIAHWLACIWYAIGNMEQPHMDSRIGWLHNLGDQIGKPYNSS
GLGGPSIKDKYVTALYFTFSSLTSVGFGNVSPNTNSEKIFSICVMLIGSLMYASIFGNVS
AIIQRLYSGTARYHTQMLRVREFIRFHQIPNPLRQRLEEYFQHAWSYTNGIDMNAVLKGF
PECLQADICLHLNRSLLQHCKPFRGATKGCLRALAMKFKTTHAPPGDTLVHAGDLLTALY
FISRGSIEILRGDVVVAILGKNDIFGEPLNLYARPGKSNGDVRALTYCDLHKIHRDDLLE
VLDMYPEFSDHFWSSLEITFNLRDTNMIPGSPGSTELEGGFSRQRKRKLSFRRRTDKDTE
QPGEVSALGPGRAGAGPSSRGRPGGPWGESPSSGPSSPESSEDEGPGRSSSPLRLVPFSS
PRPPGEPPGGEPLMEDCEKSSDTCNPLSGAFSGVSNIFSFWGDSRGRQYQELPRCPAPTP
SLLNIPLSSPGRRPRGDVESRLDALQRQLNRLETRLSADMATVLQLLQRQMTLVPPAYSA
VTTPGPGPTSTSPLLPVSPLPTLTLDSLSQVSQFMACEELPPGAPELPQEGPTRRLSLPG
QLGALTSQPLHRHGSDPGS
|
|
|
|---|
| BDBM50028854 |
|---|
| n/a |
|---|
| Name | BDBM50028854 |
|---|
| Synonyms: | CHEMBL3342693 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C23H22F5N5O2 |
|---|
| Mol. Mass. | 495.4451 |
|---|
| SMILES | CC(C)C(=O)NCc1cnc(C(F)F)c(c1)C(=O)Nc1nc(C)c([nH]1)-c1ccc(cc1)C(F)(F)F |
|---|
| Structure | 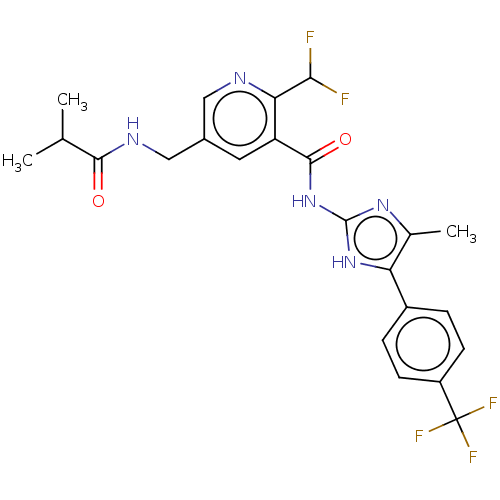
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Schiffler, MA; Antonysamy, S; Bhattachar, SN; Campanale, KM; Chandrasekhar, S; Condon, B; Desai, PV; Fisher, MJ; Groshong, C; Harvey, A; Hickey, MJ; Hughes, NE; Jones, SA; Kim, EJ; Kuklish, SL; Luz, JG; Norman, BH; Rathmell, RE; Rizzo, JR; Seng, TW; Thibodeaux, SJ; Woods, TA; York, JS; Yu, XP Discovery and Characterization of 2-Acylaminoimidazole Microsomal Prostaglandin E Synthase-1 Inhibitors. J Med Chem59:194-205 (2016) [PubMed] Article
Schiffler, MA; Antonysamy, S; Bhattachar, SN; Campanale, KM; Chandrasekhar, S; Condon, B; Desai, PV; Fisher, MJ; Groshong, C; Harvey, A; Hickey, MJ; Hughes, NE; Jones, SA; Kim, EJ; Kuklish, SL; Luz, JG; Norman, BH; Rathmell, RE; Rizzo, JR; Seng, TW; Thibodeaux, SJ; Woods, TA; York, JS; Yu, XP Discovery and Characterization of 2-Acylaminoimidazole Microsomal Prostaglandin E Synthase-1 Inhibitors. J Med Chem59:194-205 (2016) [PubMed] Article