| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Complement factor D |
|---|
| Ligand | BDBM50236213 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1658835 (CHEMBL4008447) |
|---|
| Kd | 500000±n/a nM |
|---|
| Citation |  Vulpetti, A; Randl, S; Rüdisser, S; Ostermann, N; Erbel, P; Mac Sweeney, A; Zoller, T; Salem, B; Gerhartz, B; Cumin, F; Hommel, U; Dalvit, C; Lorthiois, E; Maibaum, J Structure-Based Library Design and Fragment Screening for the Identification of Reversible Complement Factor D Protease Inhibitors. J Med Chem60:1946-1958 (2017) [PubMed] Article Vulpetti, A; Randl, S; Rüdisser, S; Ostermann, N; Erbel, P; Mac Sweeney, A; Zoller, T; Salem, B; Gerhartz, B; Cumin, F; Hommel, U; Dalvit, C; Lorthiois, E; Maibaum, J Structure-Based Library Design and Fragment Screening for the Identification of Reversible Complement Factor D Protease Inhibitors. J Med Chem60:1946-1958 (2017) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Complement factor D |
|---|
| Name: | Complement factor D |
|---|
| Synonyms: | Adipsin | C3 convertase activator | CFAD_HUMAN | CFD | DF | PFD | Properdin factor D |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 27039.19 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P00746 |
|---|
| Residue: | 253 |
|---|
| Sequence: | MHSWERLAVLVLLGAAACAAPPRGRILGGREAEAHARPYMASVQLNGAHLCGGVLVAEQW
VLSAAHCLEDAADGKVQVLLGAHSLSQPEPSKRLYDVLRAVPHPDSQPDTIDHDLLLLQL
SEKATLGPAVRPLPWQRVDRDVAPGTLCDVAGWGIVNHAGRRPDSLQHVLLPVLDRATCN
RRTHHDGAITERLMCAESNRRDSCKGDSGGPLVCGGVLEGVVTSGSRVCGNRKKPGIYTR
VASYAAWIDSVLA
|
|
|
|---|
| BDBM50236213 |
|---|
| n/a |
|---|
| Name | BDBM50236213 |
|---|
| Synonyms: | 2-(3-Benzylureido)Benzoic Acid | CHEMBL561499 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C15H14N2O3 |
|---|
| Mol. Mass. | 270.2833 |
|---|
| SMILES | OC(=O)c1ccccc1NC(=O)NCc1ccccc1 |
|---|
| Structure | 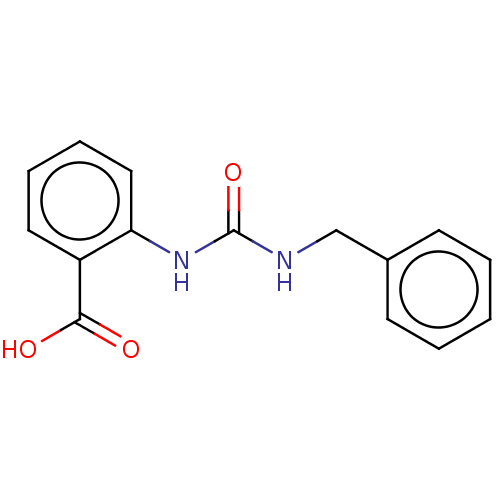
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Vulpetti, A; Randl, S; Rüdisser, S; Ostermann, N; Erbel, P; Mac Sweeney, A; Zoller, T; Salem, B; Gerhartz, B; Cumin, F; Hommel, U; Dalvit, C; Lorthiois, E; Maibaum, J Structure-Based Library Design and Fragment Screening for the Identification of Reversible Complement Factor D Protease Inhibitors. J Med Chem60:1946-1958 (2017) [PubMed] Article
Vulpetti, A; Randl, S; Rüdisser, S; Ostermann, N; Erbel, P; Mac Sweeney, A; Zoller, T; Salem, B; Gerhartz, B; Cumin, F; Hommel, U; Dalvit, C; Lorthiois, E; Maibaum, J Structure-Based Library Design and Fragment Screening for the Identification of Reversible Complement Factor D Protease Inhibitors. J Med Chem60:1946-1958 (2017) [PubMed] Article