| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Glutamate receptor 3 |
|---|
| Ligand | BDBM50004863 |
|---|
| Substrate/Competitor | n/a |
|---|
| Ki | >10000±n/a nM |
|---|
| Comments | PDSP_96 |
|---|
| Citation |  Wright, RA; Arnold, MB; Wheeler, WJ; Ornstein, PL; Schoepp, DD [3H]LY341495 binding to group II metabotropic glutamate receptors in rat brain. J Pharmacol Exp Ther298:453-60 (2001) [PubMed] Wright, RA; Arnold, MB; Wheeler, WJ; Ornstein, PL; Schoepp, DD [3H]LY341495 binding to group II metabotropic glutamate receptors in rat brain. J Pharmacol Exp Ther298:453-60 (2001) [PubMed] |
|---|
| More Info.: | Get all data from this article |
|---|
| |
| Glutamate receptor 3 |
|---|
| Name: | Glutamate receptor 3 |
|---|
| Synonyms: | GRIA3_RAT | Glur3 | Glutamate receptor 3 | Glutamate receptor ionotropic, AMPA 3 | Glutamate-AMPA | Gria3 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 100383.57 |
|---|
| Organism: | RAT |
|---|
| Description: | Glutamate-AMPA GRIA3 RAT::P19492 |
|---|
| Residue: | 888 |
|---|
| Sequence: | MGQSVLRAVFFLVLGLLGHSHGGFPNTISIGGLFMRNTVQEHSAFRFAVQLYNTNQNTTE
KPFHLNYHVDHLDSSNSFSVTNAFCSQFSRGVYAIFGFYDQMSMNTLTSFCGALHTSFVT
PSFPTDADVQFVIQMRPALKGAILSLLSYYKWEKFVYLYDTERGFSVLQAIMEAAVQNNW
QVTARSVGNIKDVQEFRRIIEEMDRRQEKRYLIDCEVERINTILEQVVILGKHSRGYHYM
LANLGFTDILLERVMHGGANITGFQIVNNENPMVQQFIQRWVRLDEREFPEAKNAPLKYT
SALTHDAILVIAEAFRYLRRQRVDVSRRGSAGDCLANPAVPWSQGIDIERALKMVQVQGM
TGNIQFDTYGRRTNYTIDVYEMKVSGSRKAGYWNEYERFVPFSDQQISNDSSSSENRTIV
VTTILESPYVMYKKNHEQLEGNERYEGYCVDLAYEIAKHVRIKYKLSIVGDGKYGARDPE
TKIWNGMVGELVYGRADIAVAPLTITLVREEVIDFSKPFMSLGISIMIKKPQKSKPGVFS
FLDPLAYEIWMCIVFAYIGVSVVLFLVSRFSPYEWHLEDNNEEPRDPQSPPDPPNEFGIF
NSLWFSLGAFMQQGCDISPRSLSGRIVGGVWWFFTLIIISSYTANLAAFLTVERMVSPIE
SAEDLAKQTEIAYGTLDSGSTKEFFRRSKIAVYEKMWSYMKSAEPSVFTKTTADGVARVR
KSKGKFAFLLESTMNEYIEQRKPCDTMKVGGNLDSKGYGVATPKGSALGNAVNLAVLKLN
EQGLLDKLKNKWWYDKGECGSGGGDSKDKTSALSLSNVAGVFYILVGGLGLAMMVALIEF
CYKSRAESKRMKLTKNTQNFKPAPATNTQNYATYREGYNVYGTESVKI
|
|
|
|---|
| BDBM50004863 |
|---|
| n/a |
|---|
| Name | BDBM50004863 |
|---|
| Synonyms: | (+/-)-trans-ACPD | (1S,3S)-1-Amino-cyclopentane-1,3-dicarboxylic acid | (1S,3S)-1-aminocyclopentane-1,3-dicarboxylic acid | 1-Amino-cyclopentane-1,3-dicarboxylic acid (APCD) | 1-Amino-cyclopentane-1,3-dicarboxylic acid(cis-(1S,3S)-ACPD) | 1R,3R-ACPD | 1R,3S-ACPD | 1S, 3R-ACPD | CHEMBL29726 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C7H11NO4 |
|---|
| Mol. Mass. | 173.1665 |
|---|
| SMILES | N[C@]1(CC[C@@H](C1)C(O)=O)C(O)=O |
|---|
| Structure | 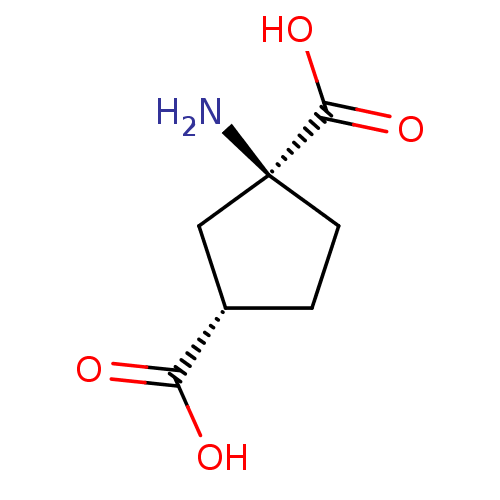
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Wright, RA; Arnold, MB; Wheeler, WJ; Ornstein, PL; Schoepp, DD [3H]LY341495 binding to group II metabotropic glutamate receptors in rat brain. J Pharmacol Exp Ther298:453-60 (2001) [PubMed]
Wright, RA; Arnold, MB; Wheeler, WJ; Ornstein, PL; Schoepp, DD [3H]LY341495 binding to group II metabotropic glutamate receptors in rat brain. J Pharmacol Exp Ther298:453-60 (2001) [PubMed]