

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Neurotensin receptor type 2 | ||
| Ligand | BDBM50240339 | ||
| Substrate/Competitor | n/a | ||
| Ki | 3.8±n/a nM | ||
| Comments | PDSP_3751 | ||
| Citation |  Pettibone, DJ; Hess, JF; Hey, PJ; Jacobson, MA; Leviten, M; Lis, EV; Mallorga, PJ; Pascarella, DM; Snyder, MA; Williams, JB; Zeng, Z The effects of deleting the mouse neurotensin receptor NTR1 on central and peripheral responses to neurotensin. J Pharmacol Exp Ther300:305-13 (2002) [PubMed] Article Pettibone, DJ; Hess, JF; Hey, PJ; Jacobson, MA; Leviten, M; Lis, EV; Mallorga, PJ; Pascarella, DM; Snyder, MA; Williams, JB; Zeng, Z The effects of deleting the mouse neurotensin receptor NTR1 on central and peripheral responses to neurotensin. J Pharmacol Exp Ther300:305-13 (2002) [PubMed] Article | ||
| More Info.: | Get all data from this article | ||
| Neurotensin receptor type 2 | |||
| Name: | Neurotensin receptor type 2 | ||
| Synonyms: | NTR2_MOUSE | Neurotensin 2 | Neurotensin receptor type 2 | Ntsr2 | ||
| Type: | Enzyme Catalytic Domain | ||
| Mol. Mass.: | 46391.18 | ||
| Organism: | MOUSE | ||
| Description: | P70310 | ||
| Residue: | 416 | ||
| Sequence: |
| ||
| BDBM50240339 | |||
| n/a | |||
| Name | BDBM50240339 | ||
| Synonyms: | (S)-2-((S)-2-((S)-2-((S)-1-((S)-6-amino-2-((S)-5-guanidino-2-(methylamino)pentanamido)hexanoyl)pyrrolidine-2-carboxamido)-3-(1H-indol-3-yl)propanamido)-3,3-dimethylbutanamido)-4-methylpentanoic acid | 2-{(R)-(S)-2-[(S)-2-({(S)-1-[(S)-6-Amino-2-((S)-5-guanidino-2-methylamino-pentanoylamino)-hexanoyl]-pyrrolidine-2-carbonyl}-amino)-3-(1H-indol-3-yl)-propionylamino]-3,3-dimethyl-butyrylamino}-4-methyl-pentanoic acid | CHEMBL266571 | Neurotensin 2 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C41H67N11O7 | ||
| Mol. Mass. | 826.0402 | ||
| SMILES | CN[C@@H](CCCN=C(N)N)C(=O)N[C@@H](CCCCN)C(=O)N1CCC[C@H]1C(=O)N[C@@H](Cc1c[nH]c2ccccc12)C(=O)N[C@H](C(=O)N[C@@H](CC(C)C)C(O)=O)C(C)(C)C |r,wU:2.1,43.45,wD:25.26,29.29,47.49,13.12,(-9.76,-17.57,;-8.43,-18.35,;-7.09,-17.6,;-7.09,-16.06,;-8.41,-15.28,;-8.4,-13.73,;-9.73,-12.95,;-9.72,-11.41,;-11.06,-10.63,;-8.38,-10.64,;-5.76,-18.38,;-5.77,-19.91,;-4.42,-17.61,;-3.09,-18.39,;-3.1,-19.93,;-4.44,-20.7,;-4.45,-22.24,;-5.78,-23,;-5.8,-24.54,;-1.74,-17.63,;-1.73,-16.1,;-.42,-18.41,;-0,-19.94,;1.55,-19.94,;2.08,-18.51,;.87,-17.56,;.94,-16,;-.36,-15.18,;2.3,-15.29,;3.6,-16.11,;3.53,-17.65,;4.83,-18.49,;6.26,-17.92,;7.23,-19.11,;6.4,-20.41,;6.81,-21.89,;5.72,-22.98,;4.24,-22.59,;3.84,-21.1,;4.92,-20.02,;4.97,-15.4,;5.05,-13.86,;6.27,-16.23,;7.64,-15.53,;8.93,-16.36,;8.86,-17.9,;10.3,-15.65,;11.59,-16.49,;11.52,-18.02,;12.82,-18.86,;12.82,-20.4,;14.19,-18.15,;12.96,-15.78,;14.25,-16.61,;13.04,-14.25,;7.72,-13.99,;7.71,-12.44,;9.26,-14.02,;6.18,-13.96,)| | ||
| Structure | 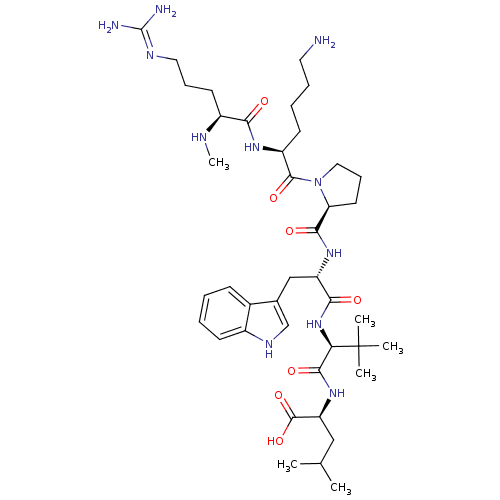
| ||