| Reaction Details |
|---|
 | Report a problem with these data |
| Target | AMP deaminase 1 |
|---|
| Ligand | BDBM154582 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | AMPD Enzymatic Activity Assay |
|---|
| pH | 7.4±n/a |
|---|
| IC50 | 4.5e+3±n/a nM |
|---|
| Comments | extracted |
|---|
| Citation |  Admyre, T; Amrot-Fors, L; Andersson, M; Bauer, M; Bjursell, M; Drmota, T; Hallen, S; Hartleib-Geschwindner, J; Lindmark, B; Liu, J; Löfgren, L; Rohman, M; Selmi, N; Wallenius, K Inhibition of AMP deaminase activity does not improve glucose control in rodent models of insulin resistance or diabetes. Chem Biol21:1486-96 (2014) [PubMed] Article Admyre, T; Amrot-Fors, L; Andersson, M; Bauer, M; Bjursell, M; Drmota, T; Hallen, S; Hartleib-Geschwindner, J; Lindmark, B; Liu, J; Löfgren, L; Rohman, M; Selmi, N; Wallenius, K Inhibition of AMP deaminase activity does not improve glucose control in rodent models of insulin resistance or diabetes. Chem Biol21:1486-96 (2014) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| AMP deaminase 1 |
|---|
| Name: | AMP deaminase 1 |
|---|
| Synonyms: | AMP deaminase 1 (mAMPD1) | AMPD1_MOUSE | Ampd1 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 86110.42 |
|---|
| Organism: | Mus musculus (Mouse) |
|---|
| Description: | n/a |
|---|
| Residue: | 745 |
|---|
| Sequence: | MPLFKLTGQGKQIDDAMRSFAEKVFASEVKDEGGRHEISPFDVDEICPISLHEMQAHIFH
MENLSMDGRRKRRFQGRKTVNLSIPQSETSSTKLSHIEEFISSSPTYESVPDFQRVQITG
DYASGVTVEDFEVVCKGLYRALCIREKYMQKSFQRFPKTPSKYLRNIDGEALVGNESFYP
VFTPPPKKGEDPFRTEDLPANLGYHLKMKAGVIYIYPDEAAANRDEPKPYPYPNLDDFLD
DMNFLLALIAQGPVKTYAHRRLKFLSSKFQVHQMLNEMDELKELKNNPHRDFYNCRKVDT
HIHAAACMNQKHLLRFIKKSYHIDADRVVYSTKEKSLTLKELFAKLNMHPYDLTVDSLDV
HAGRQTFQRFDKFNDKYNPVGASELRDLYLKTDNYINGEYFATIIKEVGADLVEAKYQHA
EPRLSIYGRSPDEWNKLSSWFVCNRIYCPNMTWMIQVPRIYDVFRSKNFLPHFGKMLENI
FLPVFEATINPQAHPDLSVFLKHITGFDSVDDESKHSGHMFSSKSPKPEEWTMENNPSYT
YYAYYMYANITVLNSLRKERGMNTFLFRPHCGEAGALTHLMTAFMIADNISHGLNLKKSP
VLQYLFFLAQIPIAMSPLSNNSLFLEYAKNPFLDFLQKGLMISLSTDDPMQFHFTKEPLM
EEYAIAAQVFKLSTCDMCEVARNSVLQCGISHEEKAKFLGNNYLEEGPVGNDIRRTNVAQ
IRMAYRYETWCYELNLIAEGLKATE
|
|
|
|---|
| BDBM154582 |
|---|
| n/a |
|---|
| Name | BDBM154582 |
|---|
| Synonyms: | 6-[4-({[(1R)-1-(isoquinolin-8-yl)ethyl]amino}methyl)phenyl]pyridine-3-carboxylic acid (Compound 2) |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C24H21N3O2 |
|---|
| Mol. Mass. | 383.4424 |
|---|
| SMILES | C[C@@H](NCc1ccc(cc1)-c1ccc(cn1)C(O)=O)c1cccc2ccncc12 |r| |
|---|
| Structure | 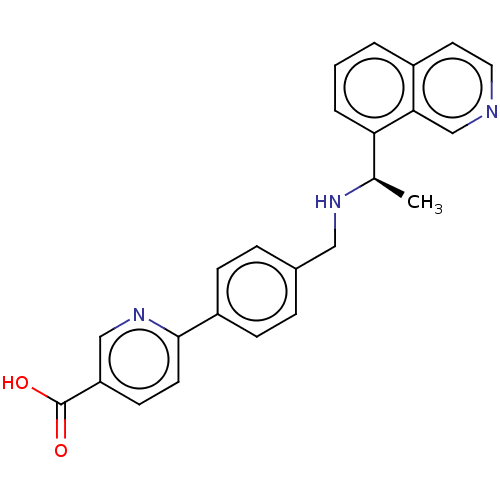
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Admyre, T; Amrot-Fors, L; Andersson, M; Bauer, M; Bjursell, M; Drmota, T; Hallen, S; Hartleib-Geschwindner, J; Lindmark, B; Liu, J; Löfgren, L; Rohman, M; Selmi, N; Wallenius, K Inhibition of AMP deaminase activity does not improve glucose control in rodent models of insulin resistance or diabetes. Chem Biol21:1486-96 (2014) [PubMed] Article
Admyre, T; Amrot-Fors, L; Andersson, M; Bauer, M; Bjursell, M; Drmota, T; Hallen, S; Hartleib-Geschwindner, J; Lindmark, B; Liu, J; Löfgren, L; Rohman, M; Selmi, N; Wallenius, K Inhibition of AMP deaminase activity does not improve glucose control in rodent models of insulin resistance or diabetes. Chem Biol21:1486-96 (2014) [PubMed] Article