

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Cellular tumor antigen p53 [18-26]/E3 ubiquitin-protein ligase Mdm2 [2-188] | ||
| Ligand | BDBM236464 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | Time Resolved Fluorescence Energy Transfer (TR-FRET) Assay | ||
| Temperature | 298.15±n/a K | ||
| IC50 | 0.134±n/a nM | ||
| Comments | extracted | ||
| Citation |  Cotesta, S; Furet, P; Guagnano, V; Holzer, P; Kallen, J; Mah, R; Masuya, K; Schlapbach, A; Stutz, S; Vaupel, A Pyrrolopyrrolidinone compounds US Patent US9365576 Publication Date 6/14/2016 Cotesta, S; Furet, P; Guagnano, V; Holzer, P; Kallen, J; Mah, R; Masuya, K; Schlapbach, A; Stutz, S; Vaupel, A Pyrrolopyrrolidinone compounds US Patent US9365576 Publication Date 6/14/2016 | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Cellular tumor antigen p53 [18-26]/E3 ubiquitin-protein ligase Mdm2 [2-188] | |||
| Name: | Cellular tumor antigen p53 [18-26]/E3 ubiquitin-protein ligase Mdm2 [2-188] | ||
| Synonyms: | p53-MDM2 | ||
| Type: | Protein | ||
| Mol. Mass.: | n/a | ||
| Description: | n/a | ||
| Components: | This complex has 2 components. | ||
| Component 1 | |||
| Name: | Cellular tumor antigen p53 [18-26] | ||
| Synonyms: | P53 | P53_HUMAN | TP53 | Tumor suppressor p53 | ||
| Type: | Protein | ||
| Mol. Mass.: | 1122.31 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | aa 18-26 | ||
| Residue: | 9 | ||
| Sequence: |
| ||
| Component 2 | |||
| Name: | E3 ubiquitin-protein ligase Mdm2 [2-188] | ||
| Synonyms: | E3 ubiquitin-protein ligase Mdm2 | MDM2 | MDM2_HUMAN | ||
| Type: | Protein | ||
| Mol. Mass.: | 21187.98 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | aa 2-188 | ||
| Residue: | 187 | ||
| Sequence: |
| ||
| BDBM236464 | |||
| n/a | |||
| Name | BDBM236464 | ||
| Synonyms: | US9365576, 220 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C30H29Cl2N5O6 | ||
| Mol. Mass. | 626.487 | ||
| SMILES | CCOC(=O)c1c2C(=O)N(C(c2n(C(C)C)c1-c1cnc(OC)nc1OC)c1ccc(Cl)cc1)c1cc(Cl)c(=O)n(C)c1 |(3.26,6.8,;2.17,5.71,;2.57,4.22,;1.48,3.13,;.39,4.22,;1.08,1.64,;-.38,1.17,;-1.85,1.64,;-2.62,2.98,;-2.75,.4,;-1.85,-.85,;-.38,-.37,;1.08,-.85,;1.48,-2.33,;.39,-3.42,;2.97,-2.73,;1.98,.4,;3.52,.4,;4.29,-.94,;5.83,-.94,;6.6,.4,;8.14,.4,;8.91,-.94,;5.83,1.73,;4.29,1.73,;3.52,3.07,;4.29,4.4,;-2.25,-2.33,;-3.74,-2.73,;-4.13,-4.22,;-3.05,-5.31,;-3.44,-6.8,;-1.56,-4.91,;-1.16,-3.42,;-4.29,.4,;-5.06,1.73,;-6.6,1.73,;-7.37,3.07,;-7.37,.4,;-8.91,.4,;-6.6,-.94,;-7.37,-2.27,;-5.06,-.94,)| | ||
| Structure | 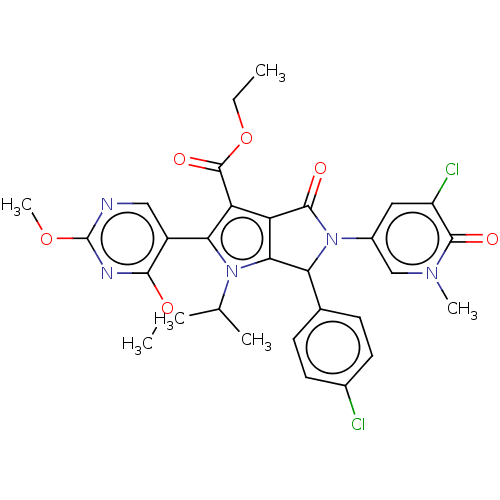
| ||