

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Bile acid receptor | ||
| Ligand | BDBM30331 | ||
| Substrate/Competitor | BDBM10852 | ||
| Meas. Tech. | FXR Transient Transfection Assay | ||
| EC50 | 500±n/a nM | ||
| Comments | 90% efficacy. | ||
| Citation |  Akwabi-Ameyaw, A; Bass, JY; Caldwell, RD; Caravella, JA; Chen, L; Creech, KL; Deaton, DN; Jones, SA; Kaldor, I; Liu, Y; Madauss, KP; Marr, HB; McFadyen, RB; Miller, AB; Iii, FN; Parks, DJ; Spearing, PK; Todd, D; Williams, SP; Wisely, GB Conformationally constrained farnesoid X receptor (FXR) agonists: Naphthoic acid-based analogs of GW 4064. Bioorg Med Chem Lett18:4339-43 (2008) [PubMed] Article Akwabi-Ameyaw, A; Bass, JY; Caldwell, RD; Caravella, JA; Chen, L; Creech, KL; Deaton, DN; Jones, SA; Kaldor, I; Liu, Y; Madauss, KP; Marr, HB; McFadyen, RB; Miller, AB; Iii, FN; Parks, DJ; Spearing, PK; Todd, D; Williams, SP; Wisely, GB Conformationally constrained farnesoid X receptor (FXR) agonists: Naphthoic acid-based analogs of GW 4064. Bioorg Med Chem Lett18:4339-43 (2008) [PubMed] Article | ||
| More Info.: | Get all data from this article, Solution Info, Assay Method | ||
| Bile acid receptor | |||
| Name: | Bile acid receptor | ||
| Synonyms: | BAR | Bile acid receptor FXR | FXR | Farnesol receptor HRR-1 | HRR1 | NR1H4 | NR1H4_HUMAN | Nuclear receptor subfamily 1 group H member 4 | RIP14 | RXR-interacting protein 14 | Retinoid X receptor-interacting protein 14 | farnesoid x receptor | ||
| Type: | Nuclear Receptor | ||
| Mol. Mass.: | 55916.24 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q96RI1 | ||
| Residue: | 486 | ||
| Sequence: |
| ||
| BDBM30331 | |||
| BDBM10852 | |||
| Name | BDBM30331 | ||
| Synonyms: | Naphthoic acid-based analog, 1d | ||
| Type | Small organic molecule | ||
| Emp. Form. | C30H23Cl2NO4 | ||
| Mol. Mass. | 532.414 | ||
| SMILES | CC(C)c1onc(c1COc1ccc(cc1)-c1ccc2ccc(cc2c1)C(O)=O)-c1c(Cl)cccc1Cl |(-3.6,6.71,;-2.16,7.25,;-1.91,8.77,;-.97,6.27,;.52,6.66,;1.35,5.37,;.38,4.18,;-1.06,4.74,;-2.39,3.97,;-3.73,4.74,;-5.06,3.97,;-6.39,4.74,;-7.73,3.97,;-7.73,2.43,;-6.39,1.66,;-5.06,2.43,;-9.06,1.66,;-9.06,.12,;-10.39,-.65,;-11.73,.12,;-13.06,-.65,;-14.39,.12,;-14.39,1.66,;-13.06,2.43,;-11.73,1.66,;-10.39,2.43,;-15.73,2.43,;-17.06,1.66,;-15.73,3.97,;.76,2.69,;-.28,1.55,;-1.78,1.89,;.18,.08,;1.68,-.26,;2.73,.88,;2.27,2.35,;3.31,3.48,)| | ||
| Structure | 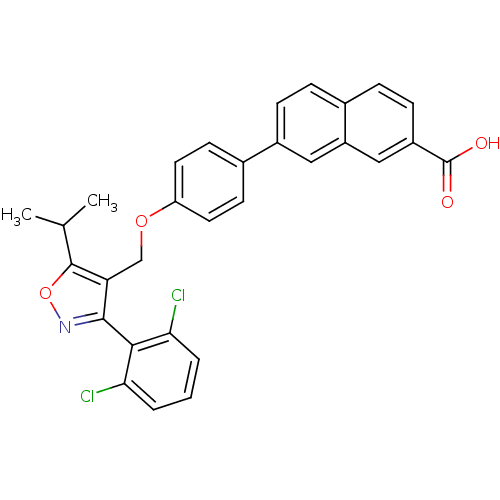
| ||