| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Mitogen-activated protein kinase 1 |
|---|
| Ligand | BDBM50313013 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1672659 (CHEMBL4022688) |
|---|
| IC50 | >10000±n/a nM |
|---|
| Citation |  Wagman, AS; Boyce, RS; Brown, SP; Fang, E; Goff, D; Jansen, JM; Le, VP; Levine, BH; Ng, SC; Ni, ZJ; Nuss, JM; Pfister, KB; Ramurthy, S; Renhowe, PA; Ring, DB; Shu, W; Subramanian, S; Zhou, XA; Shafer, CM; Harrison, SD; Johnson, KW; Bussiere, DE Synthesis, Binding Mode, and Antihyperglycemic Activity of Potent and Selective (5-Imidazol-2-yl-4-phenylpyrimidin-2-yl)[2-(2-pyridylamino)ethyl]amine Inhibitors of Glycogen Synthase Kinase 3. J Med Chem60:8482-8514 (2017) [PubMed] Article Wagman, AS; Boyce, RS; Brown, SP; Fang, E; Goff, D; Jansen, JM; Le, VP; Levine, BH; Ng, SC; Ni, ZJ; Nuss, JM; Pfister, KB; Ramurthy, S; Renhowe, PA; Ring, DB; Shu, W; Subramanian, S; Zhou, XA; Shafer, CM; Harrison, SD; Johnson, KW; Bussiere, DE Synthesis, Binding Mode, and Antihyperglycemic Activity of Potent and Selective (5-Imidazol-2-yl-4-phenylpyrimidin-2-yl)[2-(2-pyridylamino)ethyl]amine Inhibitors of Glycogen Synthase Kinase 3. J Med Chem60:8482-8514 (2017) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Mitogen-activated protein kinase 1 |
|---|
| Name: | Mitogen-activated protein kinase 1 |
|---|
| Synonyms: | ERK2 | ERT1 | Extracellular signal-regulated kinase 2 | Extracellular signal-regulated kinase 2 (ERK-2) | Extracellular signal-regulated kinase 2 (ERK2) | MAP Kinase 2/ERK2 | MAPK 2 | MAPK1 | MK01_HUMAN | Mitogen activated kinase 1 (ERK-2) | Mitogen-activated protein kinase 1 (ERK-2) | Mitogen-activated protein kinase 1 (ERK2) | Mitogen-activated protein kinase 2 | PRKM1 | PRKM2 | p42-MAPK |
|---|
| Type: | Ser/Thr Protein Kinase |
|---|
| Mol. Mass.: | 41392.76 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P28482 |
|---|
| Residue: | 360 |
|---|
| Sequence: | MAAAAAAGAGPEMVRGQVFDVGPRYTNLSYIGEGAYGMVCSAYDNVNKVRVAIKKISPFE
HQTYCQRTLREIKILLRFRHENIIGINDIIRAPTIEQMKDVYIVQDLMETDLYKLLKTQH
LSNDHICYFLYQILRGLKYIHSANVLHRDLKPSNLLLNTTCDLKICDFGLARVADPDHDH
TGFLTEYVATRWYRAPEIMLNSKGYTKSIDIWSVGCILAEMLSNRPIFPGKHYLDQLNHI
LGILGSPSQEDLNCIINLKARNYLLSLPHKNKVPWNRLFPNADSKALDLLDKMLTFNPHK
RIEVEQALAHPYLEQYYDPSDEPIAEAPFKFDMELDDLPKEKLKELIFEETARFQPGYRS
|
|
|
|---|
| BDBM50313013 |
|---|
| n/a |
|---|
| Name | BDBM50313013 |
|---|
| Synonyms: | CHEMBL1080901 | CT-98024 | N2-(2-(4-(2,4-dichlorophenyl)-5-(1H-imidazol-2-yl)pyrimidin-2-ylamino)ethyl)-5-nitropyridine-2,6-diamine |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H17Cl2N9O2 |
|---|
| Mol. Mass. | 486.314 |
|---|
| SMILES | Nc1nc(NCCNc2ncc(-c3ncc[nH]3)c(n2)-c2ccc(Cl)cc2Cl)ccc1[N+]([O-])=O |
|---|
| Structure | 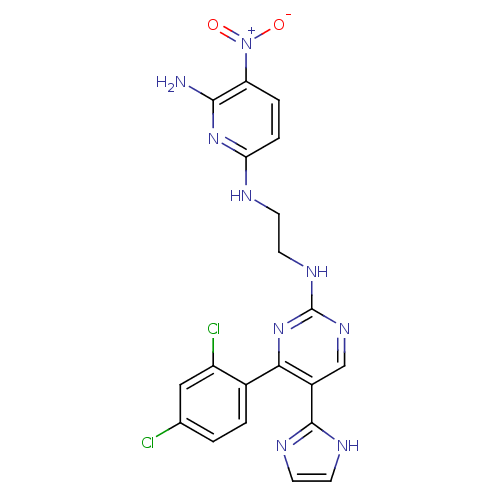
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Wagman, AS; Boyce, RS; Brown, SP; Fang, E; Goff, D; Jansen, JM; Le, VP; Levine, BH; Ng, SC; Ni, ZJ; Nuss, JM; Pfister, KB; Ramurthy, S; Renhowe, PA; Ring, DB; Shu, W; Subramanian, S; Zhou, XA; Shafer, CM; Harrison, SD; Johnson, KW; Bussiere, DE Synthesis, Binding Mode, and Antihyperglycemic Activity of Potent and Selective (5-Imidazol-2-yl-4-phenylpyrimidin-2-yl)[2-(2-pyridylamino)ethyl]amine Inhibitors of Glycogen Synthase Kinase 3. J Med Chem60:8482-8514 (2017) [PubMed] Article
Wagman, AS; Boyce, RS; Brown, SP; Fang, E; Goff, D; Jansen, JM; Le, VP; Levine, BH; Ng, SC; Ni, ZJ; Nuss, JM; Pfister, KB; Ramurthy, S; Renhowe, PA; Ring, DB; Shu, W; Subramanian, S; Zhou, XA; Shafer, CM; Harrison, SD; Johnson, KW; Bussiere, DE Synthesis, Binding Mode, and Antihyperglycemic Activity of Potent and Selective (5-Imidazol-2-yl-4-phenylpyrimidin-2-yl)[2-(2-pyridylamino)ethyl]amine Inhibitors of Glycogen Synthase Kinase 3. J Med Chem60:8482-8514 (2017) [PubMed] Article