

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | E3 ubiquitin-protein ligase Mdm2 | ||
| Ligand | BDBM129723 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1795643 (CHEMBL4267760) | ||
| IC50 | 0.170000±n/a nM | ||
| Citation |  Vaupel, A; Holzer, P; Ferretti, S; Guagnano, V; Kallen, J; Mah, R; Masuya, K; Ruetz, S; Rynn, C; Schlapbach, A; Stachyra, T; Stutz, S; Todorov, M; Jeay, S; Furet, P In vitro and in vivo characterization of a novel, highly potent p53-MDM2 inhibitor. Bioorg Med Chem Lett28:3404-3408 (2018) [PubMed] Article Vaupel, A; Holzer, P; Ferretti, S; Guagnano, V; Kallen, J; Mah, R; Masuya, K; Ruetz, S; Rynn, C; Schlapbach, A; Stachyra, T; Stutz, S; Todorov, M; Jeay, S; Furet, P In vitro and in vivo characterization of a novel, highly potent p53-MDM2 inhibitor. Bioorg Med Chem Lett28:3404-3408 (2018) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| E3 ubiquitin-protein ligase Mdm2 | |||
| Name: | E3 ubiquitin-protein ligase Mdm2 | ||
| Synonyms: | Double minute 2 protein | Double minute 2 protein (HDM2) | E3 ubiquitin-protein ligase Mdm2 (p53-binding protein Mdm2) | Hdm2 | Human Double Minute 2 (HDM2) | MDM2 | MDM2-MDMX | MDM2_HUMAN | p53-Binding Protein MDM2 | p53-binding protein | ||
| Type: | Oncoprotein | ||
| Mol. Mass.: | 55196.54 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q00987 | ||
| Residue: | 491 | ||
| Sequence: |
| ||
| BDBM129723 | |||
| n/a | |||
| Name | BDBM129723 | ||
| Synonyms: | US8815926, 2 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C29H26Cl2FN3O2 | ||
| Mol. Mass. | 538.44 | ||
| SMILES | COc1cccc(F)c1-c1nc2C(=O)N(C(c2n1C(C)C)c1ccc(Cl)cc1C)c1cc(Cl)ccc1C |(4.68,5.6,;3.91,4.26,;4.68,2.93,;6.22,2.93,;6.99,1.6,;6.22,.26,;4.68,.26,;3.91,-1.07,;3.91,1.6,;2.37,1.6,;1.46,2.84,;,2.37,;-1.46,2.84,;-2.23,4.18,;-2.37,1.6,;-1.46,.35,;,.83,;1.46,.35,;1.86,-1.14,;.77,-2.22,;3.35,-1.53,;-1.86,-1.14,;-.77,-2.22,;-1.17,-3.71,;-2.66,-4.11,;-3.06,-5.6,;-3.75,-3.02,;-3.35,-1.53,;-4.44,-.45,;-3.91,1.6,;-4.68,.26,;-6.22,.26,;-6.99,-1.07,;-6.99,1.6,;-6.22,2.93,;-4.68,2.93,;-3.91,4.26,)| | ||
| Structure | 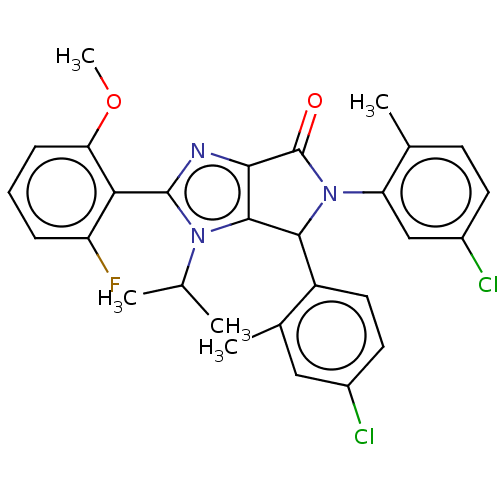
| ||