| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Uridine phosphorylase 1 |
|---|
| Ligand | BDBM50031017 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_212951 (CHEMBL873996) |
|---|
| IC50 | 24000±n/a nM |
|---|
| Citation |  Orr, GF; Musso, DL; Boswell, GE; Kelley, JL; Joyner, SS; Davis, ST; Baccanari, DP Inhibition of uridine phosphorylase: synthesis and structure-activity relationships of aryl-substituted 5-benzyluracils and 1-[(2-hydroxyethoxy)methyl]-5-benzyluracils. J Med Chem38:3850-6 (1995) [PubMed] Orr, GF; Musso, DL; Boswell, GE; Kelley, JL; Joyner, SS; Davis, ST; Baccanari, DP Inhibition of uridine phosphorylase: synthesis and structure-activity relationships of aryl-substituted 5-benzyluracils and 1-[(2-hydroxyethoxy)methyl]-5-benzyluracils. J Med Chem38:3850-6 (1995) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Uridine phosphorylase 1 |
|---|
| Name: | Uridine phosphorylase 1 |
|---|
| Synonyms: | UPP1_MOUSE | Up | Upp | Upp1 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 34086.29 |
|---|
| Organism: | Mus musculus |
|---|
| Description: | ChEMBL_212951 |
|---|
| Residue: | 311 |
|---|
| Sequence: | MAATGTEAKDLENHHNDCFIQLSNPNIAAMKEDVLYHFNLSTSTHDFPAMFGDVKFVCVG
GSSSRMNTFIKYVAAELGLDHPGKEYPNICAGTDRYAMYKAGPVLSVSHGMGIPSIGIML
HELIKMLYHARCSNITIIRIGTSGGIGLEPGSVVITQQAVNECFKPEFEQIVLGKRVIRN
TNLDAQLVQELVQCSSDLNEFPMVVGNTMCTLDFYEGQGRLDGALCSYTEKDKQSYLRAA
HAAGVRNIEMESSVFATMCSACGLKAAVVCVTLLDRLQGDQINTPHDVLVEYQQRPQRLV
GHFIKKSLGRA
|
|
|
|---|
| BDBM50031017 |
|---|
| n/a |
|---|
| Name | BDBM50031017 |
|---|
| Synonyms: | 5-(2-Chloro-benzyl)-1H-pyrimidine-2,4-dione | CHEMBL127529 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C11H9ClN2O2 |
|---|
| Mol. Mass. | 236.654 |
|---|
| SMILES | Clc1ccccc1Cc1c[nH]c(=O)[nH]c1=O |
|---|
| Structure | 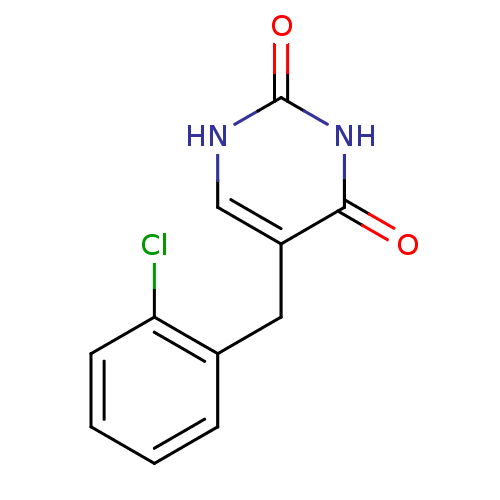
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Orr, GF; Musso, DL; Boswell, GE; Kelley, JL; Joyner, SS; Davis, ST; Baccanari, DP Inhibition of uridine phosphorylase: synthesis and structure-activity relationships of aryl-substituted 5-benzyluracils and 1-[(2-hydroxyethoxy)methyl]-5-benzyluracils. J Med Chem38:3850-6 (1995) [PubMed]
Orr, GF; Musso, DL; Boswell, GE; Kelley, JL; Joyner, SS; Davis, ST; Baccanari, DP Inhibition of uridine phosphorylase: synthesis and structure-activity relationships of aryl-substituted 5-benzyluracils and 1-[(2-hydroxyethoxy)methyl]-5-benzyluracils. J Med Chem38:3850-6 (1995) [PubMed]