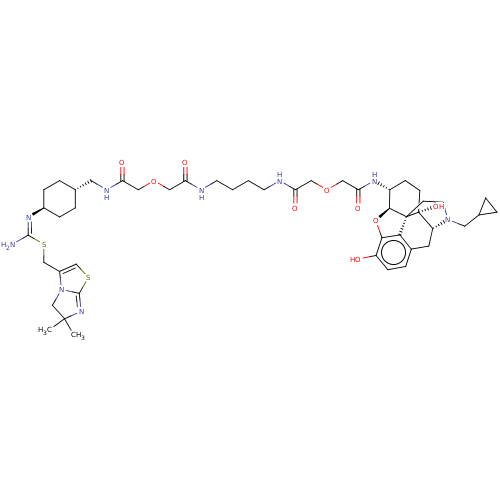
| SMILES | [H][C@@]12Oc3c4c(C[C@@]5([H])N(CC6CC6)CC[C@@]14[C@@]5(O)CC[C@H]2NC(=O)COCC(=O)NCCCCNC(=O)COCC(=O)NC[C@H]1CC[C@@H](CC1)\N=C(\N)SCC1=CSC2=NC(C)(C)CN12)ccc3O |r,wU:16.16,17.21,21.26,1.0,48.56,wD:7.7,45.49,t:62,65,THB:10:9:17:4.5.6,(10.99,-22.68,;9.28,-22.68,;10.2,-24.25,;9.27,-25.68,;7.98,-24.94,;6.66,-25.68,;5.34,-24.94,;5.34,-23.46,;4.38,-22.47,;3.82,-23.41,;2.47,-22.64,;2.46,-21.12,;3.2,-19.79,;1.67,-19.8,;4.79,-24.2,;6.67,-24.2,;7.98,-23.41,;6.67,-22.65,;5.32,-21.88,;6.67,-21.12,;7.99,-20.36,;9.3,-21.15,;10.65,-20.39,;10.67,-18.85,;9.34,-18.06,;12.01,-18.09,;13.34,-18.87,;14.68,-18.12,;16.01,-18.9,;15.99,-20.44,;17.35,-18.14,;18.68,-18.93,;20.02,-18.17,;21.35,-18.95,;22.7,-18.2,;24.02,-18.98,;25.37,-18.22,;25.38,-16.68,;26.69,-19.01,;28.04,-18.25,;29.36,-19.03,;30.71,-18.27,;30.72,-16.73,;32.04,-19.06,;33.38,-18.3,;34.71,-19.09,;34.69,-20.63,;36.01,-21.41,;37.35,-20.66,;37.37,-19.11,;36.04,-18.32,;38.68,-21.45,;40.02,-20.69,;41.35,-21.48,;40.05,-19.15,;41.39,-18.4,;41.41,-16.86,;42.67,-15.97,;42.21,-14.49,;40.67,-14.48,;39.44,-13.56,;38.18,-14.45,;36.68,-14.84,;37.09,-13.35,;38.64,-15.91,;40.17,-15.93,;6.66,-27.21,;7.98,-27.98,;9.27,-27.21,;10.63,-28,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Ma, H; Wang, H; Li, M; Barreto-de-Souza, V; Reinecke, BA; Gunta, R; Zheng, Y; Kang, G; Nassehi, N; Zhang, H; An, J; Selley, DE; Hauser, KF; Zhang, Y Bivalent Ligand Aiming Putative Mu Opioid Receptor and Chemokine Receptor CXCR4 Dimers in Opioid Enhanced HIV-1 Entry. ACS Med Chem Lett11:2318-2324 (2020) [PubMed] Article
Ma, H; Wang, H; Li, M; Barreto-de-Souza, V; Reinecke, BA; Gunta, R; Zheng, Y; Kang, G; Nassehi, N; Zhang, H; An, J; Selley, DE; Hauser, KF; Zhang, Y Bivalent Ligand Aiming Putative Mu Opioid Receptor and Chemokine Receptor CXCR4 Dimers in Opioid Enhanced HIV-1 Entry. ACS Med Chem Lett11:2318-2324 (2020) [PubMed] Article