| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Protein mono-ADP-ribosyltransferase PARP3 |
|---|
| Ligand | BDBM50593627 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2268422 |
|---|
| IC50 | 110±n/a nM |
|---|
| Citation |  Buchstaller, HP; Anlauf, U; Dorsch, D; Kögler, S; Kuhn, D; Lehmann, M; Leuthner, B; Lodholz, S; Musil, D; Radtki, D; Rettig, C; Ritzert, C; Rohdich, F; Schneider, R; Wegener, A; Weigt, S; Wilkinson, K; Esdar, C Optimization of a Screening Hit toward M2912, an Oral Tankyrase Inhibitor with Antitumor Activity in Colorectal Cancer Models. J Med Chem64:10371-10392 (2021) [PubMed] Buchstaller, HP; Anlauf, U; Dorsch, D; Kögler, S; Kuhn, D; Lehmann, M; Leuthner, B; Lodholz, S; Musil, D; Radtki, D; Rettig, C; Ritzert, C; Rohdich, F; Schneider, R; Wegener, A; Weigt, S; Wilkinson, K; Esdar, C Optimization of a Screening Hit toward M2912, an Oral Tankyrase Inhibitor with Antitumor Activity in Colorectal Cancer Models. J Med Chem64:10371-10392 (2021) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Protein mono-ADP-ribosyltransferase PARP3 |
|---|
| Name: | Protein mono-ADP-ribosyltransferase PARP3 |
|---|
| Synonyms: | ADP-ribosyltransferase diphtheria toxin-like 3 | ADPRT-3 | ADPRT3 | ADPRTL3 | ARTD3 | IRT1 | NAD(+) ADP-ribosyltransferase 3 | PARP-3 | PARP3 | PARP3_HUMAN | Poly [ADP-ribose] polymerase 3 | Poly[ADP-ribose] synthase 3 | hPARP-3 | pADPRT-3 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 60091.40 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_109545 |
|---|
| Residue: | 533 |
|---|
| Sequence: | MAPKPKPWVQTEGPEKKKGRQAGREEDPFRSTAEALKAIPAEKRIIRVDPTCPLSSNPGT
QVYEDYNCTLNQTNIENNNNKFYIIQLLQDSNRFFTCWNRWGRVGEVGQSKINHFTRLED
AKKDFEKKFREKTKNNWAERDHFVSHPGKYTLIEVQAEDEAQEAVVKVDRGPVRTVTKRV
QPCSLDPATQKLITNIFSKEMFKNTMALMDLDVKKMPLGKLSKQQIARGFEALEALEEAL
KGPTDGGQSLEELSSHFYTVIPHNFGHSQPPPINSPELLQAKKDMLLVLADIELAQALQA
VSEQEKTVEEVPHPLDRDYQLLKCQLQLLDSGAPEYKVIQTYLEQTGSNHRCPTLQHIWK
VNQEGEEDRFQAHSKLGNRKLLWHGTNMAVVAAILTSGLRIMPHSGGRVGKGIYFASENS
KSAGYVIGMKCGAHHVGYMFLGEVALGREHHINTDNPSLKSPPPGFDSVIARGHTEPDPT
QDTELELDGQQVVVPQGQPVPCPEFSSSTFSQSEYLIYQESQCRLRYLLEVHL
|
|
|
|---|
| BDBM50593627 |
|---|
| n/a |
|---|
| Name | BDBM50593627 |
|---|
| Synonyms: | CHEMBL5199558 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C17H18N2O2 |
|---|
| Mol. Mass. | 282.337 |
|---|
| SMILES | Cc1ccc2n1cc([nH]c2=O)-c1ccc(cc1)C(C)(C)O |
|---|
| Structure | 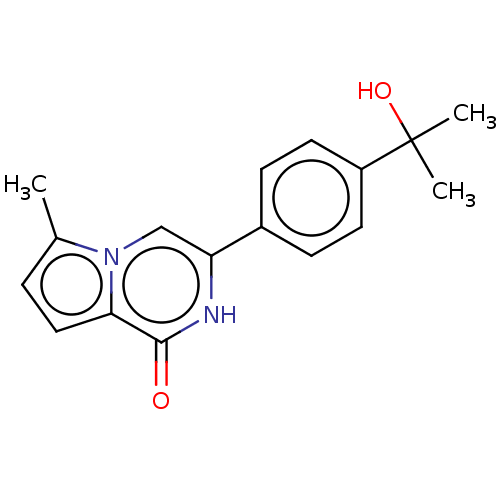
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Buchstaller, HP; Anlauf, U; Dorsch, D; Kögler, S; Kuhn, D; Lehmann, M; Leuthner, B; Lodholz, S; Musil, D; Radtki, D; Rettig, C; Ritzert, C; Rohdich, F; Schneider, R; Wegener, A; Weigt, S; Wilkinson, K; Esdar, C Optimization of a Screening Hit toward M2912, an Oral Tankyrase Inhibitor with Antitumor Activity in Colorectal Cancer Models. J Med Chem64:10371-10392 (2021) [PubMed]
Buchstaller, HP; Anlauf, U; Dorsch, D; Kögler, S; Kuhn, D; Lehmann, M; Leuthner, B; Lodholz, S; Musil, D; Radtki, D; Rettig, C; Ritzert, C; Rohdich, F; Schneider, R; Wegener, A; Weigt, S; Wilkinson, K; Esdar, C Optimization of a Screening Hit toward M2912, an Oral Tankyrase Inhibitor with Antitumor Activity in Colorectal Cancer Models. J Med Chem64:10371-10392 (2021) [PubMed]