| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Poly [ADP-ribose] polymerase 1 |
|---|
| Ligand | BDBM50335641 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_714646 (CHEMBL1663324) |
|---|
| IC50 | 60±n/a nM |
|---|
| Citation |  Pierre, F; Chua, PC; O'Brien, SE; Siddiqui-Jain, A; Bourbon, P; Haddach, M; Michaux, J; Nagasawa, J; Schwaebe, MK; Stefan, E; Vialettes, A; Whitten, JP; Chen, TK; Darjania, L; Stansfield, R; Anderes, K; Bliesath, J; Drygin, D; Ho, C; Omori, M; Proffitt, C; Streiner, N; Trent, K; Rice, WG; Ryckman, DM Discovery and SAR of 5-(3-chlorophenylamino)benzo[c][2,6]naphthyridine-8-carboxylic acid (CX-4945), the first clinical stage inhibitor of protein kinase CK2 for the treatment of cancer. J Med Chem54:635-54 (2011) [PubMed] Article Pierre, F; Chua, PC; O'Brien, SE; Siddiqui-Jain, A; Bourbon, P; Haddach, M; Michaux, J; Nagasawa, J; Schwaebe, MK; Stefan, E; Vialettes, A; Whitten, JP; Chen, TK; Darjania, L; Stansfield, R; Anderes, K; Bliesath, J; Drygin, D; Ho, C; Omori, M; Proffitt, C; Streiner, N; Trent, K; Rice, WG; Ryckman, DM Discovery and SAR of 5-(3-chlorophenylamino)benzo[c][2,6]naphthyridine-8-carboxylic acid (CX-4945), the first clinical stage inhibitor of protein kinase CK2 for the treatment of cancer. J Med Chem54:635-54 (2011) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Poly [ADP-ribose] polymerase 1 |
|---|
| Name: | Poly [ADP-ribose] polymerase 1 |
|---|
| Synonyms: | (ARTD1 or PARP1) | 2.4.2.- | 2.4.2.30 | ADP-ribosyltransferase diphtheria toxin-like 1 | ADPRT | ADPRT 1 | ARTD1 | DNA ADP-ribosyltransferase PARP1 | Human diphtheria toxin-like ADP-ribosyltransferase (ARTD1 or PARP1) | NAD(+) ADP-ribosyltransferase 1 | NT-PARP-1 | PARP-1 | PARP1 | PARP1_HUMAN | PPOL | Poly [ADP-ribose] polymerase (PARP) | Poly [ADP-ribose] polymerase 1 (PARP) | Poly [ADP-ribose] polymerase 1 (PARP-1) | Poly [ADP-ribose] polymerase 1 (PARP1) | Poly [ADP-ribose] polymerase 1, 24-kDa form | Poly [ADP-ribose] polymerase 1, 28-kDa form | Poly [ADP-ribose] polymerase 1, 89-kDa form | Poly [ADP-ribose] polymerase 1, processed C-terminus | Poly [ADP-ribose] polymerase 1, processed N-terminus | Poly [ADP-ribose] polymerase-1 | Poly(ADP-ribose) polymerase 1 (PARP1) | Poly(ADP-ribose) polymerase-1 (ARTD1/PARP1) | Poly[ADP-ribose] synthase 1 | Protein poly-ADP-ribosyltransferase PARP1 | Synonyms=ADPRT |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | 113114.22 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P09874 |
|---|
| Residue: | 1014 |
|---|
| Sequence: | MAESSDKLYRVEYAKSGRASCKKCSESIPKDSLRMAIMVQSPMFDGKVPHWYHFSCFWKV
GHSIRHPDVEVDGFSELRWDDQQKVKKTAEAGGVTGKGQDGIGSKAEKTLGDFAAEYAKS
NRSTCKGCMEKIEKGQVRLSKKMVDPEKPQLGMIDRWYHPGCFVKNREELGFRPEYSASQ
LKGFSLLATEDKEALKKQLPGVKSEGKRKGDEVDGVDEVAKKKSKKEKDKDSKLEKALKA
QNDLIWNIKDELKKVCSTNDLKELLIFNKQQVPSGESAILDRVADGMVFGALLPCEECSG
QLVFKSDAYYCTGDVTAWTKCMVKTQTPNRKEWVTPKEFREISYLKKLKVKKQDRIFPPE
TSASVAATPPPSTASAPAAVNSSASADKPLSNMKILTLGKLSRNKDEVKAMIEKLGGKLT
GTANKASLCISTKKEVEKMNKKMEEVKEANIRVVSEDFLQDVSASTKSLQELFLAHILSP
WGAEVKAEPVEVVAPRGKSGAALSKKSKGQVKEEGINKSEKRMKLTLKGGAAVDPDSGLE
HSAHVLEKGGKVFSATLGLVDIVKGTNSYYKLQLLEDDKENRYWIFRSWGRVGTVIGSNK
LEQMPSKEDAIEHFMKLYEEKTGNAWHSKNFTKYPKKFYPLEIDYGQDEEAVKKLTVNPG
TKSKLPKPVQDLIKMIFDVESMKKAMVEYEIDLQKMPLGKLSKRQIQAAYSILSEVQQAV
SQGSSDSQILDLSNRFYTLIPHDFGMKKPPLLNNADSVQAKVEMLDNLLDIEVAYSLLRG
GSDDSSKDPIDVNYEKLKTDIKVVDRDSEEAEIIRKYVKNTHATTHNAYDLEVIDIFKIE
REGECQRYKPFKQLHNRRLLWHGSRTTNFAGILSQGLRIAPPEAPVTGYMFGKGIYFADM
VSKSANYCHTSQGDPIGLILLGEVALGNMYELKHASHISKLPKGKHSVKGLGKTTPDPSA
NISLDGVDVPLGTGISSGVNDTSLLYNEYIVYDIAQVNLKYLLKLKFNFKTSLW
|
|
|
|---|
| BDBM50335641 |
|---|
| n/a |
|---|
| Name | BDBM50335641 |
|---|
| Synonyms: | 4-Oxo-4,5-dihydrothieno[3,2-c]quinoline-7-carboxamide | CHEMBL1652712 | US8168651, Compound TABLE 15.45 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C12H8N2O2S |
|---|
| Mol. Mass. | 244.269 |
|---|
| SMILES | NC(=O)c1ccc2c3sccc3c(=O)[nH]c2c1 |
|---|
| Structure | 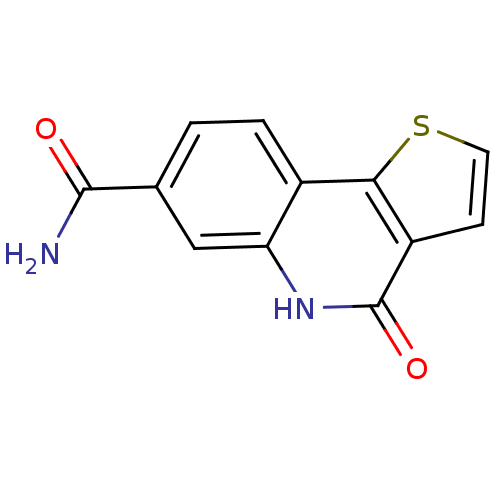
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Pierre, F; Chua, PC; O'Brien, SE; Siddiqui-Jain, A; Bourbon, P; Haddach, M; Michaux, J; Nagasawa, J; Schwaebe, MK; Stefan, E; Vialettes, A; Whitten, JP; Chen, TK; Darjania, L; Stansfield, R; Anderes, K; Bliesath, J; Drygin, D; Ho, C; Omori, M; Proffitt, C; Streiner, N; Trent, K; Rice, WG; Ryckman, DM Discovery and SAR of 5-(3-chlorophenylamino)benzo[c][2,6]naphthyridine-8-carboxylic acid (CX-4945), the first clinical stage inhibitor of protein kinase CK2 for the treatment of cancer. J Med Chem54:635-54 (2011) [PubMed] Article
Pierre, F; Chua, PC; O'Brien, SE; Siddiqui-Jain, A; Bourbon, P; Haddach, M; Michaux, J; Nagasawa, J; Schwaebe, MK; Stefan, E; Vialettes, A; Whitten, JP; Chen, TK; Darjania, L; Stansfield, R; Anderes, K; Bliesath, J; Drygin, D; Ho, C; Omori, M; Proffitt, C; Streiner, N; Trent, K; Rice, WG; Ryckman, DM Discovery and SAR of 5-(3-chlorophenylamino)benzo[c][2,6]naphthyridine-8-carboxylic acid (CX-4945), the first clinical stage inhibitor of protein kinase CK2 for the treatment of cancer. J Med Chem54:635-54 (2011) [PubMed] Article