| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Methionine aminopeptidase 1 |
|---|
| Ligand | BDBM50129647 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_105134 (CHEMBL715018) |
|---|
| IC50 | >100000±n/a nM |
|---|
| Citation |  Luo, QL; Li, JY; Liu, ZY; Chen, LL; Li, J; Qian, Z; Shen, Q; Li, Y; Lushington, GH; Ye, QZ; Nan, FJ Discovery and structural modification of inhibitors of methionine aminopeptidases from Escherichia coli and Saccharomyces cerevisiae. J Med Chem46:2631-40 (2003) [PubMed] Article Luo, QL; Li, JY; Liu, ZY; Chen, LL; Li, J; Qian, Z; Shen, Q; Li, Y; Lushington, GH; Ye, QZ; Nan, FJ Discovery and structural modification of inhibitors of methionine aminopeptidases from Escherichia coli and Saccharomyces cerevisiae. J Med Chem46:2631-40 (2003) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Methionine aminopeptidase 1 |
|---|
| Name: | Methionine aminopeptidase 1 |
|---|
| Synonyms: | MAP 1 | MAP1 | MAP1_YEAST | MetAP 1 | Peptidase M 1 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 43382.92 |
|---|
| Organism: | Saccharomyces cerevisiae |
|---|
| Description: | ChEMBL_10701 |
|---|
| Residue: | 387 |
|---|
| Sequence: | MSTATTTVTTSDQASHPTKIYCSGLQCGRETSSQMKCPVCLKQGIVSIFCDTSCYENNYK
AHKALHNAKDGLEGAYDPFPKFKYSGKVKASYPLTPRRYVPEDIPKPDWAANGLPVSEQR
NDRLNNIPIYKKDQIKKIRKACMLGREVLDIAAAHVRPGITTDELDEIVHNETIKRGAYP
SPLNYYNFPKSLCTSVNEVICHGVPDKTVLKEGDIVNLDVSLYYQGYHADLNETYYVGEN
ISKEALNTTETSRECLKLAIKMCKPGTTFQELGDHIEKHATENKCSVVRTYCGHGVGEFF
HCSPNIPHYAKNRTPGVMKPGMVFTIEPMINEGTWKDMTWPDDWTSTTQDGKLSAQFEHT
LLVTEHGVEILTARNKKSPGGPRQRIK
|
|
|
|---|
| BDBM50129647 |
|---|
| n/a |
|---|
| Name | BDBM50129647 |
|---|
| Synonyms: | 3-(4-Fluoro-benzoylamino)-pyridine-2-carboxylic acid thiazol-2-ylamide | CHEMBL329320 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C16H11FN4O2S |
|---|
| Mol. Mass. | 342.348 |
|---|
| SMILES | Fc1ccc(cc1)C(=O)Nc1cccnc1C(=O)Nc1nccs1 |
|---|
| Structure | 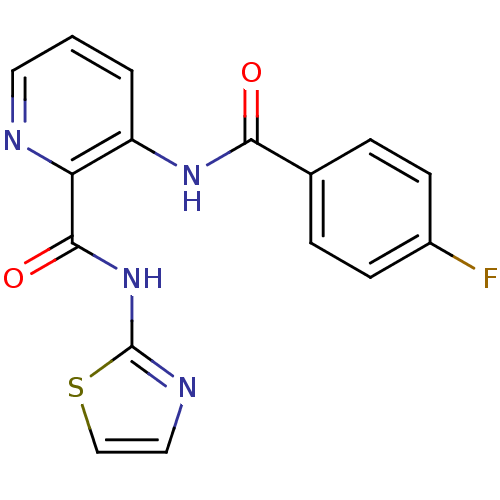
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Luo, QL; Li, JY; Liu, ZY; Chen, LL; Li, J; Qian, Z; Shen, Q; Li, Y; Lushington, GH; Ye, QZ; Nan, FJ Discovery and structural modification of inhibitors of methionine aminopeptidases from Escherichia coli and Saccharomyces cerevisiae. J Med Chem46:2631-40 (2003) [PubMed] Article
Luo, QL; Li, JY; Liu, ZY; Chen, LL; Li, J; Qian, Z; Shen, Q; Li, Y; Lushington, GH; Ye, QZ; Nan, FJ Discovery and structural modification of inhibitors of methionine aminopeptidases from Escherichia coli and Saccharomyces cerevisiae. J Med Chem46:2631-40 (2003) [PubMed] Article