| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Muscarinic acetylcholine receptor M1 |
|---|
| Ligand | BDBM50423967 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_935580 (CHEMBL2320116) |
|---|
| EC50 | >30000±n/a nM |
|---|
| Citation |  Le, U; Melancon, BJ; Bridges, TM; Vinson, PN; Utley, TJ; Lamsal, A; Rodriguez, AL; Venable, D; Sheffler, DJ; Jones, CK; Blobaum, AL; Wood, MR; Daniels, JS; Conn, PJ; Niswender, CM; Lindsley, CW; Hopkins, CR Discovery of a selective M4 positive allosteric modulator based on the 3-amino-thieno[2,3-b]pyridine-2-carboxamide scaffold: development of ML253, a potent and brain penetrant compound that is active in a preclinical model of schizophrenia. Bioorg Med Chem Lett23:346-50 (2012) [PubMed] Article Le, U; Melancon, BJ; Bridges, TM; Vinson, PN; Utley, TJ; Lamsal, A; Rodriguez, AL; Venable, D; Sheffler, DJ; Jones, CK; Blobaum, AL; Wood, MR; Daniels, JS; Conn, PJ; Niswender, CM; Lindsley, CW; Hopkins, CR Discovery of a selective M4 positive allosteric modulator based on the 3-amino-thieno[2,3-b]pyridine-2-carboxamide scaffold: development of ML253, a potent and brain penetrant compound that is active in a preclinical model of schizophrenia. Bioorg Med Chem Lett23:346-50 (2012) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Muscarinic acetylcholine receptor M1 |
|---|
| Name: | Muscarinic acetylcholine receptor M1 |
|---|
| Synonyms: | ACM1_RAT | Cholinergic, muscarinic M1 | Chrm-1 | Chrm1 | cholinergic receptor, muscarinic 1 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 51390.46 |
|---|
| Organism: | RAT |
|---|
| Description: | P08482 |
|---|
| Residue: | 460 |
|---|
| Sequence: | MNTSVPPAVSPNITVLAPGKGPWQVAFIGITTGLLSLATVTGNLLVLISFKVNTELKTVN
NYFLLSLACADLIIGTFSMNLYTTYLLMGHWALGTLACDLWLALDYVASNASVMNLLLIS
FDRYFSVTRPLSYRAKRTPRRAALMIGLAWLVSFVLWAPAILFWQYLVGERTVLAGQCYI
QFLSQPIITFGTAMAAFYLPVTVMCTLYWRIYRETENRARELAALQGSETPGKGGGSSSS
SERSQPGAEGSPESPPGRCCRCCRAPRLLQAYSWKEEEEEDEGSMESLTSSEGEEPGSEV
VIKMPMVDSEAQAPTKQPPKSSPNTVKRPTKKGRDRGGKGQKPRGKEQLAKRKTFSLVKE
KKAARTLSAILLAFILTWTPYNIMVLVSTFCKDCVPETLWELGYWLCYVNSTVNPMCYAL
CNKAFRDTFRLLLLCRWDKRRWRKIPKRPGSVHRTPSRQC
|
|
|
|---|
| BDBM50423967 |
|---|
| n/a |
|---|
| Name | BDBM50423967 |
|---|
| Synonyms: | CHEMBL2057417 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C16H15ClN4OS |
|---|
| Mol. Mass. | 346.835 |
|---|
| SMILES | Cc1nc2sc(C(=O)NCc3ccncc3)c(N)c2c(C)c1Cl |
|---|
| Structure | 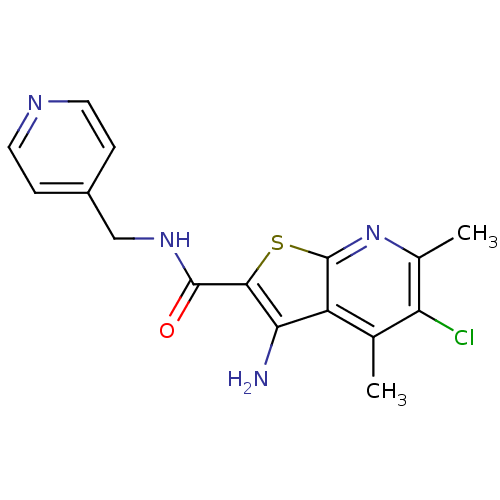
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Le, U; Melancon, BJ; Bridges, TM; Vinson, PN; Utley, TJ; Lamsal, A; Rodriguez, AL; Venable, D; Sheffler, DJ; Jones, CK; Blobaum, AL; Wood, MR; Daniels, JS; Conn, PJ; Niswender, CM; Lindsley, CW; Hopkins, CR Discovery of a selective M4 positive allosteric modulator based on the 3-amino-thieno[2,3-b]pyridine-2-carboxamide scaffold: development of ML253, a potent and brain penetrant compound that is active in a preclinical model of schizophrenia. Bioorg Med Chem Lett23:346-50 (2012) [PubMed] Article
Le, U; Melancon, BJ; Bridges, TM; Vinson, PN; Utley, TJ; Lamsal, A; Rodriguez, AL; Venable, D; Sheffler, DJ; Jones, CK; Blobaum, AL; Wood, MR; Daniels, JS; Conn, PJ; Niswender, CM; Lindsley, CW; Hopkins, CR Discovery of a selective M4 positive allosteric modulator based on the 3-amino-thieno[2,3-b]pyridine-2-carboxamide scaffold: development of ML253, a potent and brain penetrant compound that is active in a preclinical model of schizophrenia. Bioorg Med Chem Lett23:346-50 (2012) [PubMed] Article