

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Cyclin-dependent kinase 5 activator 1 | ||
| Ligand | BDBM50261213 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1695674 (CHEMBL4046564) | ||
| IC50 | 71±n/a nM | ||
| Citation |  Park, SJ; Kim, E; Yoo, M; Lee, JY; Park, CH; Hwang, JY; Ha, JD Synthesis and biological evaluation of N9-cis-cyclobutylpurine derivatives for use as cyclin-dependent kinase (CDK) inhibitors. Bioorg Med Chem Lett27:4399-4404 (2017) [PubMed] Article Park, SJ; Kim, E; Yoo, M; Lee, JY; Park, CH; Hwang, JY; Ha, JD Synthesis and biological evaluation of N9-cis-cyclobutylpurine derivatives for use as cyclin-dependent kinase (CDK) inhibitors. Bioorg Med Chem Lett27:4399-4404 (2017) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Cyclin-dependent kinase 5 activator 1 | |||
| Name: | Cyclin-dependent kinase 5 activator 1 | ||
| Synonyms: | CDK5/p35 | Cyclin-Dependent Kinase 5 (CDK5) | Cyclin-dependent kinase 5 regulatory subunit 1 | Cyclin-dependent kinase 5/CDK5 activator 1 | ||
| Type: | Protein Complex | ||
| Mol. Mass.: | n/a | ||
| Description: | n/a | ||
| Components: | This complex has 2 components. | ||
| Component 1 | |||
| Name: | Cyclin-dependent kinase 5 | ||
| Synonyms: | CDK5 | CDK5_HUMAN | CDKN5 | Cell division protein kinase 5 | Cyclin-dependent kinase 5 (CDK5/ p25) | Cyclin-dependent kinase 5 (CDK5/p35) | Cyclin-dependent-like kinase 5 | Cyclin-dependent-like kinase 5 (CDK5) | PSSALRE | Serine/threonine-protein kinase PSSALRE | Tau protein kinase II catalytic subunit | ||
| Type: | Enzyme Subunit | ||
| Mol. Mass.: | 33308.61 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | n/a | ||
| Residue: | 292 | ||
| Sequence: |
| ||
| Component 2 | |||
| Name: | Cyclin-dependent kinase 5 activator 1 | ||
| Synonyms: | CD5R1_HUMAN | CDK5R | CDK5R1 | Cyclin-Dependent Kinase 5 Activator 1, p35 | Cyclin-dependent kinase 5 (CDK5/p25) | Cyclin-dependent kinase 5 regulatory subunit 1 | NCK5A | TPKII regulatory subunit | p35 | ||
| Type: | Enzyme Subunit | ||
| Mol. Mass.: | 34077.43 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q15078 | ||
| Residue: | 307 | ||
| Sequence: |
| ||
| BDBM50261213 | |||
| n/a | |||
| Name | BDBM50261213 | ||
| Synonyms: | CHEMBL4072013 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C22H28N8O | ||
| Mol. Mass. | 420.5107 | ||
| SMILES | Cc1cccc(n1)C(=O)N[C@H]1C[C@H](C1)n1cnc2c(NCC3CCNCC3)ncnc12 |r,wU:12.15,10.10,(35.5,-21.79,;34.42,-22.88,;32.93,-22.48,;31.84,-23.57,;32.24,-25.06,;33.72,-25.45,;34.81,-24.37,;34.13,-26.94,;35.63,-27.26,;33.1,-28.09,;31.59,-27.77,;30.29,-28.6,;29.46,-27.31,;30.75,-26.48,;27.95,-26.99,;27.32,-25.59,;25.79,-25.75,;25.47,-27.25,;24.14,-28.03,;22.81,-27.25,;21.47,-28.03,;20.14,-27.25,;20.14,-25.71,;18.81,-24.95,;17.48,-25.71,;17.48,-27.25,;18.81,-28.03,;24.14,-29.57,;25.47,-30.33,;26.81,-29.57,;26.81,-28.03,)| | ||
| Structure | 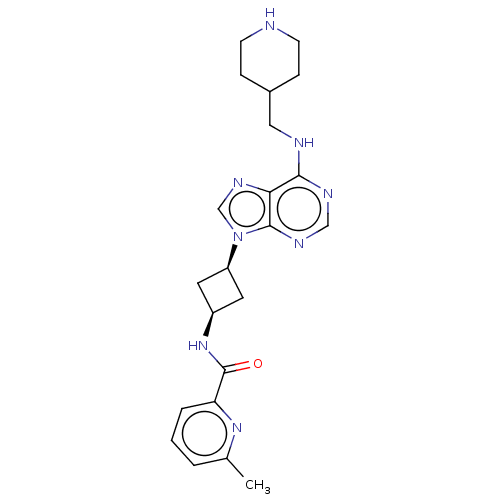
| ||