

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Cytochrome P450 2C9 | ||
| Ligand | BDBM50606313 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_2254483 (CHEMBL5168693) | ||
| IC50 | 30000±n/a nM | ||
| Citation |  Ma, C; Wu, J; Wang, L; Ji, X; Wu, Y; Miao, L; Chen, D; Zhang, L; Wu, Y; Feng, H; Tang, Y; Zhou, Q; Pei, J; Yang, X; Xu, D; You, Q; Xie, Y Discovery of Clinical Candidate NTQ1062 as a Potent and Bioavailable Akt Inhibitor for the Treatment of Human Tumors. J Med Chem65:8144-8168 (2022) [PubMed] Article Ma, C; Wu, J; Wang, L; Ji, X; Wu, Y; Miao, L; Chen, D; Zhang, L; Wu, Y; Feng, H; Tang, Y; Zhou, Q; Pei, J; Yang, X; Xu, D; You, Q; Xie, Y Discovery of Clinical Candidate NTQ1062 as a Potent and Bioavailable Akt Inhibitor for the Treatment of Human Tumors. J Med Chem65:8144-8168 (2022) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Cytochrome P450 2C9 | |||
| Name: | Cytochrome P450 2C9 | ||
| Synonyms: | (R)-limonene 6-monooxygenase | (S)-limonene 6-monooxygenase | CP2C9_HUMAN | CYP2C10 | CYP2C9 | CYPIIC9 | Cytochrome P450 2C9 (CYP2C9 ) | Cytochrome P450 2C9 (CYP2C9) | P-450MP | P450 MP-4/MP-8 | P450 PB-1 | S-mephenytoin 4-hydroxylase | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 55636.33 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P11712 | ||
| Residue: | 490 | ||
| Sequence: |
| ||
| BDBM50606313 | |||
| n/a | |||
| Name | BDBM50606313 | ||
| Synonyms: | CHEMBL5193325 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C29H37ClN6O3 | ||
| Mol. Mass. | 553.095 | ||
| SMILES | [H][C@@]12C[C@]1([H])N(CCN2C(=O)[C@H](CNC1CCC(CC1)OC)c1ccc(Cl)cc1)c1ncnc2NC(=O)C[C@@H](C)c12 |r,wU:11.13,1.0,3.4,wD:38.42,(3.08,-1.58,;2,-2.67,;3.33,-3.45,;2,-4.22,;3.48,-4.62,;.66,-5,;-.68,-4.22,;-.68,-2.67,;.66,-1.91,;.66,-.37,;1.99,.4,;-.67,.4,;-.67,1.94,;.66,2.71,;.66,4.25,;-.67,5.02,;-.67,6.56,;.66,7.33,;1.99,6.56,;1.99,5.02,;.66,8.87,;-.67,9.64,;-2.01,-.37,;-2,-1.91,;-3.34,-2.68,;-4.67,-1.92,;-6.01,-2.69,;-4.68,-.37,;-3.34,.4,;.66,-6.54,;-.68,-7.32,;-.68,-8.87,;.66,-9.64,;2,-8.87,;3.33,-9.64,;4.67,-8.87,;6.01,-9.64,;4.67,-7.32,;3.33,-6.54,;3.33,-5,;2,-7.32,)| | ||
| Structure | 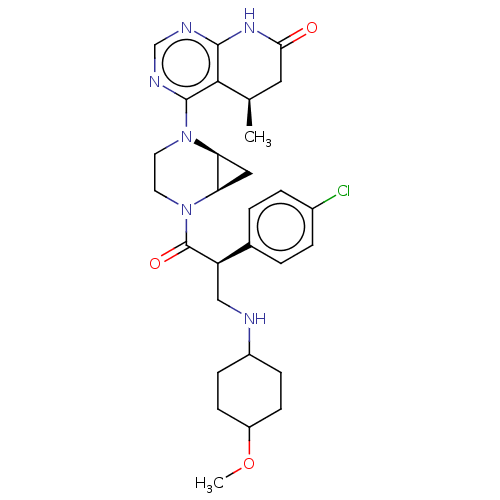
| ||