| Reaction Details |
|---|
 | Report a problem with these data |
| Target | ATP-dependent molecular chaperone HSP82 |
|---|
| Ligand | BDBM15359 |
|---|
| Substrate/Competitor | BDBM2 |
|---|
| Meas. Tech. | Colorimetric Determination of ATPase Activity |
|---|
| pH | 7.4±n/a |
|---|
| Temperature | 310.15±n/a K |
|---|
| IC50 | 13400±3000 nM |
|---|
| Citation |  Wright, L; Barril, X; Dymock, B; Sheridan, L; Surgenor, A; Beswick, M; Drysdale, M; Collier, A; Massey, A; Davies, N; Fink, A; Fromont, C; Aherne, W; Boxall, K; Sharp, S; Workman, P; Hubbard, RE Structure-activity relationships in purine-based inhibitor binding to HSP90 isoforms. Chem Biol11:775-85 (2004) [PubMed] Article Wright, L; Barril, X; Dymock, B; Sheridan, L; Surgenor, A; Beswick, M; Drysdale, M; Collier, A; Massey, A; Davies, N; Fink, A; Fromont, C; Aherne, W; Boxall, K; Sharp, S; Workman, P; Hubbard, RE Structure-activity relationships in purine-based inhibitor binding to HSP90 isoforms. Chem Biol11:775-85 (2004) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Inhibition_Run data, Solution Info, Assay Method |
|---|
| |
| ATP-dependent molecular chaperone HSP82 |
|---|
| Name: | ATP-dependent molecular chaperone HSP82 |
|---|
| Synonyms: | 82 kDa heat shock protein | ATP-dependent molecular chaperone HSP82 | HSP82 | HSP82_YEAST | HSP90 | Heat Shock Protein 90 (Hsp90) | Heat shock protein Hsp90 heat-inducible isoform |
|---|
| Type: | Molecular Chaperone |
|---|
| Mol. Mass.: | 81369.08 |
|---|
| Organism: | Saccharomyces cerevisiae |
|---|
| Description: | Histidine-tagged yeast HSP90 was transformed into E. coli and purified (>90%) by metal affinity, gel filtration, and ion-exchange chromatography. |
|---|
| Residue: | 709 |
|---|
| Sequence: | MASETFEFQAEITQLMSLIINTVYSNKEIFLRELISNASDALDKIRYKSLSDPKQLETEP
DLFIRITPKPEQKVLEIRDSGIGMTKAELINNLGTIAKSGTKAFMEALSAGADVSMIGQF
GVGFYSLFLVADRVQVISKSNDDEQYIWESNAGGSFTVTLDEVNERIGRGTILRLFLKDD
QLEYLEEKRIKEVIKRHSEFVAYPIQLVVTKEVEKEVPIPEEEKKDEEKKDEEKKDEDDK
KPKLEEVDEEEEKKPKTKKVKEEVQEIEELNKTKPLWTRNPSDITQEEYNAFYKSISNDW
EDPLYVKHFSVEGQLEFRAILFIPKRAPFDLFESKKKKNNIKLYVRRVFITDEAEDLIPE
WLSFVKGVVDSEDLPLNLSREMLQQNKIMKVIRKNIVKKLIEAFNEIAEDSEQFEKFYSA
FSKNIKLGVHEDTQNRAALAKLLRYNSTKSVDELTSLTDYVTRMPEHQKNIYYITGESLK
AVEKSPFLDALKAKNFEVLFLTDPIDEYAFTQLKEFEGKTLVDITKDFELEETDEEKAER
EKEIKEYEPLTKALKEILGDQVEKVVVSYKLLDAPAAIRTGQFGWSANMERIMKAQALRD
SSMSSYMSSKKTFEISPKSPIIKELKKRVDEGGAQDKTVKDLTKLLYETALLTSGFSLDE
PTSFASRINRLISLGLNIDEDEETETAPEASTAAPVEEVPADTEMEEVD
|
|
|
|---|
| BDBM15359 |
|---|
| BDBM2 |
|---|
| Name | BDBM15359 |
|---|
| Synonyms: | (4E,6Z,8S,9S,10E,12S,13R,14S,16R)-13-hydroxy-8,14-dimethoxy-4,10,12,16-tetramethyl-3,20,22-trioxo-19-(prop-2-en-1-ylamino)-2-azabicyclo[16.3.1]docosa-1(21),4,6,10,18-pentaen-9-yl carbamate | 17-(Allylamino)geldanamycin | 17-AAG | 17AAG | CHEMBL109480 | GLD-36 | Tanespimycin |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C31H43N3O8 |
|---|
| Mol. Mass. | 585.6884 |
|---|
| SMILES | CO[C@H]1C[C@H](C)Cc2c(O)c(NC(=O)C(C)=CC=C[C@H](OC)[C@@H](OC(N)=O)\C(C)=C\[C@H](C)[C@H]1O)cc(O)c2N=CC=C |r,w:16.16,38.39,t:28| |
|---|
| Structure | 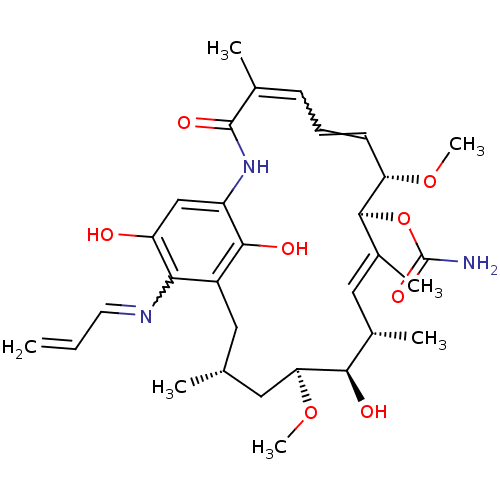
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Wright, L; Barril, X; Dymock, B; Sheridan, L; Surgenor, A; Beswick, M; Drysdale, M; Collier, A; Massey, A; Davies, N; Fink, A; Fromont, C; Aherne, W; Boxall, K; Sharp, S; Workman, P; Hubbard, RE Structure-activity relationships in purine-based inhibitor binding to HSP90 isoforms. Chem Biol11:775-85 (2004) [PubMed] Article
Wright, L; Barril, X; Dymock, B; Sheridan, L; Surgenor, A; Beswick, M; Drysdale, M; Collier, A; Massey, A; Davies, N; Fink, A; Fromont, C; Aherne, W; Boxall, K; Sharp, S; Workman, P; Hubbard, RE Structure-activity relationships in purine-based inhibitor binding to HSP90 isoforms. Chem Biol11:775-85 (2004) [PubMed] Article