| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Fatty-acid amide hydrolase 1 [30-579] |
|---|
| Ligand | BDBM26722 |
|---|
| Substrate/Competitor | BDBM22988 |
|---|
| Meas. Tech. | FAAH Carbon Filtration Assay |
|---|
| pH | 8±n/a |
|---|
| Temperature | 295.15±n/a K |
|---|
| IC50 | 16±2 nM |
|---|
| Citation |  Wang, X; Sarris, K; Kage, K; Zhang, D; Brown, SP; Kolasa, T; Surowy, C; El Kouhen, OF; Muchmore, SW; Brioni, JD; Stewart, AO Synthesis and evaluation of benzothiazole-based analogues as novel, potent, and selective fatty acid amide hydrolase inhibitors. J Med Chem52:170-80 (2009) [PubMed] Article Wang, X; Sarris, K; Kage, K; Zhang, D; Brown, SP; Kolasa, T; Surowy, C; El Kouhen, OF; Muchmore, SW; Brioni, JD; Stewart, AO Synthesis and evaluation of benzothiazole-based analogues as novel, potent, and selective fatty acid amide hydrolase inhibitors. J Med Chem52:170-80 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Fatty-acid amide hydrolase 1 [30-579] |
|---|
| Name: | Fatty-acid amide hydrolase 1 [30-579] |
|---|
| Synonyms: | Anandamide amidohydrolase 1 | FAAH1_RAT | Faah | Faah1 | Fatty Acid Amide Hydrolase | Fatty Acid Amide Hydrolic, FAAH | Fatty-acid amide hydrolase (FAAH) | Fatty-acid amide hydrolase 1 | Fatty-acid amide hydrolase 1 (FAAH) | Fatty-acid amide hydrolase 1 (aa 30-579) | Oleamide hydrolase 1 |
|---|
| Type: | Single-pass membrane protein; homodimer |
|---|
| Mol. Mass.: | 60474.00 |
|---|
| Organism: | Rattus norvegicus (rat) |
|---|
| Description: | P97612 (aa 30-579) |
|---|
| Residue: | 550 |
|---|
| Sequence: | RWTGRQKARGAATRARQKQRASLETMDKAVQRFRLQNPDLDSEALLTLPLLQLVQKLQSG
ELSPEAVFFTYLGKAWEVNKGTNCVTSYLTDCETQLSQAPRQGLLYGVPVSLKECFSYKG
HDSTLGLSLNEGMPSESDCVVVQVLKLQGAVPFVHTNVPQSMLSFDCSNPLFGQTMNPWK
SSKSPGGSSGGEGALIGSGGSPLGLGTDIGGSIRFPSAFCGICGLKPTGNRLSKSGLKGC
VYGQTAVQLSLGPMARDVESLALCLKALLCEHLFTLDPTVPPLPFREEVYRSSRPLRVGY
YETDNYTMPSPAMRRALIETKQRLEAAGHTLIPFLPNNIPYALEVLSAGGLFSDGGRSFL
QNFKGDFVDPCLGDLILILRLPSWFKRLLSLLLKPLFPRLAAFLNSMRPRSAEKLWKLQH
EIEMYRQSVIAQWKAMNLDVLLTPMLGPALDLNTPGRATGAISYTVLYNCLDFPAGVVPV
TTVTAEDDAQMELYKGYFGDIWDIILKKAMKNSVGLPVAVQCVALPWQEELCLRFMREVE
QLMTPQKQPS
|
|
|
|---|
| BDBM26722 |
|---|
| BDBM22988 |
|---|
| Name | BDBM26722 |
|---|
| Synonyms: | 4-(6-methyl-1,3-benzothiazol-2-yl)-N-{[1-(thiophene-2-sulfonyl)piperidin-4-yl]methyl}aniline | benzothiazole analogue, 15e |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C24H25N3O2S3 |
|---|
| Mol. Mass. | 483.669 |
|---|
| SMILES | Cc1ccc2nc(sc2c1)-c1ccc(NCC2CCN(CC2)S(=O)(=O)c2cccs2)cc1 |
|---|
| Structure | 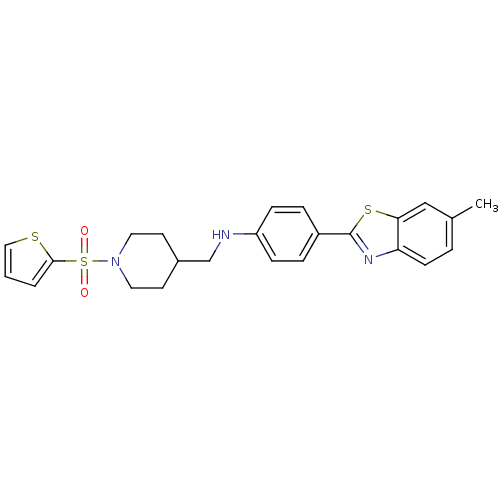
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Wang, X; Sarris, K; Kage, K; Zhang, D; Brown, SP; Kolasa, T; Surowy, C; El Kouhen, OF; Muchmore, SW; Brioni, JD; Stewart, AO Synthesis and evaluation of benzothiazole-based analogues as novel, potent, and selective fatty acid amide hydrolase inhibitors. J Med Chem52:170-80 (2009) [PubMed] Article
Wang, X; Sarris, K; Kage, K; Zhang, D; Brown, SP; Kolasa, T; Surowy, C; El Kouhen, OF; Muchmore, SW; Brioni, JD; Stewart, AO Synthesis and evaluation of benzothiazole-based analogues as novel, potent, and selective fatty acid amide hydrolase inhibitors. J Med Chem52:170-80 (2009) [PubMed] Article