| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Gag-Pol polyprotein [489-587] |
|---|
| Ligand | BDBM383 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_158031 (CHEMBL768622) |
|---|
| IC50 | 0.06±n/a nM |
|---|
| Citation |  Holloway, MK; Wai, JM; Halgren, TA; Fitzgerald, PM; Vacca, JP; Dorsey, BD; Levin, RB; Thompson, WJ; Chen, LJ; deSolms, SJ A priori prediction of activity for HIV-1 protease inhibitors employing energy minimization in the active site. J Med Chem38:305-17 (1995) [PubMed] Holloway, MK; Wai, JM; Halgren, TA; Fitzgerald, PM; Vacca, JP; Dorsey, BD; Levin, RB; Thompson, WJ; Chen, LJ; deSolms, SJ A priori prediction of activity for HIV-1 protease inhibitors employing energy minimization in the active site. J Med Chem38:305-17 (1995) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Gag-Pol polyprotein [489-587] |
|---|
| Name: | Gag-Pol polyprotein [489-587] |
|---|
| Synonyms: | Human immunodeficiency virus type 1 protease | POL_HV1H2 | Pol polyprotein | gag-pol |
|---|
| Type: | Enzyme Subunit |
|---|
| Mol. Mass.: | 10781.16 |
|---|
| Organism: | Human immunodeficiency virus type 1 |
|---|
| Description: | P04585[489-587] |
|---|
| Residue: | 99 |
|---|
| Sequence: | PQVTLWQRPLVTIKIGGQLKEALLDTGADDTVLEEMSLPGRWKPKMIGGIGGFIKVRQYD
QILIEICGHKAIGTVLVGPTPVNIIGRNLLTQIGCTLNF
|
|
|
|---|
| BDBM383 |
|---|
| n/a |
|---|
| Name | BDBM383 |
|---|
| Synonyms: | (3S)-oxolan-3-yl N-[(2S,3S,5R)-5-benzyl-3-hydroxy-5-{[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]carbamoyl}-1-phenylpentan-2-yl]carbamate | Urethane deriv. 2 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C33H38N2O6 |
|---|
| Mol. Mass. | 558.6646 |
|---|
| SMILES | O[C@@H](C[C@@H](Cc1ccccc1)C(=O)N[C@@H]1[C@H](O)Cc2ccccc12)[C@H](Cc1ccccc1)NC(=O)O[C@H]1CCOC1 |r| |
|---|
| Structure | 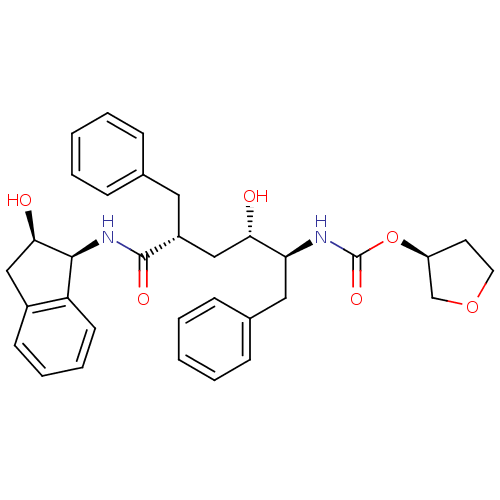
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Holloway, MK; Wai, JM; Halgren, TA; Fitzgerald, PM; Vacca, JP; Dorsey, BD; Levin, RB; Thompson, WJ; Chen, LJ; deSolms, SJ A priori prediction of activity for HIV-1 protease inhibitors employing energy minimization in the active site. J Med Chem38:305-17 (1995) [PubMed]
Holloway, MK; Wai, JM; Halgren, TA; Fitzgerald, PM; Vacca, JP; Dorsey, BD; Levin, RB; Thompson, WJ; Chen, LJ; deSolms, SJ A priori prediction of activity for HIV-1 protease inhibitors employing energy minimization in the active site. J Med Chem38:305-17 (1995) [PubMed]