| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Histone-lysine N-methyltransferase EHMT1 |
|---|
| Ligand | BDBM50300032 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1656137 (CHEMBL4005607) |
|---|
| IC50 | 13±n/a nM |
|---|
| Citation |  Xiong, Y; Li, F; Babault, N; Dong, A; Zeng, H; Wu, H; Chen, X; Arrowsmith, CH; Brown, PJ; Liu, J; Vedadi, M; Jin, J Discovery of Potent and Selective Inhibitors for G9a-Like Protein (GLP) Lysine Methyltransferase. J Med Chem60:1876-1891 (2017) [PubMed] Article Xiong, Y; Li, F; Babault, N; Dong, A; Zeng, H; Wu, H; Chen, X; Arrowsmith, CH; Brown, PJ; Liu, J; Vedadi, M; Jin, J Discovery of Potent and Selective Inhibitors for G9a-Like Protein (GLP) Lysine Methyltransferase. J Med Chem60:1876-1891 (2017) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Histone-lysine N-methyltransferase EHMT1 |
|---|
| Name: | Histone-lysine N-methyltransferase EHMT1 |
|---|
| Synonyms: | EHMT1 | EHMT1_HUMAN | EUHMTASE1 | Eu-HMTase1 | Euchromatic histone-lysine N-methyltransferase 1 | G9a-like protein 1 | GLP | GLP1 | H3-K9-HMTase 5 | Histone H3-K9 methyltransferase 5 | Histone-lysine N-methyltransferase EHMT1/EHMT2 | KIAA1876 | KMT1D | Lysine N-methyltransferase 1D |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 141443.67 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_1450367 |
|---|
| Residue: | 1298 |
|---|
| Sequence: | MAAADAEAVPARGEPQQDCCVKTELLGEETPMAADEGSAEKQAGEAHMAADGETNGSCEN
SDASSHANAAKHTQDSARVNPQDGTNTLTRIAENGVSERDSEAAKQNHVTADDFVQTSVI
GSNGYILNKPALQAQPLRTTSTLASSLPGHAAKTLPGGAGKGRTPSAFPQTPAAPPATLG
EGSADTEDRKLPAPGADVKVHRARKTMPKSVVGLHAASKDPREVREARDHKEPKEEINKN
ISDFGRQQLLPPFPSLHQSLPQNQCYMATTKSQTACLPFVLAAAVSRKKKRRMGTYSLVP
KKKTKVLKQRTVIEMFKSITHSTVGSKGEKDLGASSLHVNGESLEMDSDEDDSEELEEDD
GHGAEQAAAFPTEDSRTSKESMSEADRAQKMDGESEEEQESVDTGEEEEGGDESDLSSES
SIKKKFLKRKGKTDSPWIKPARKRRRRSRKKPSGALGSESYKSSAGSAEQTAPGDSTGYM
EVSLDSLDLRVKGILSSQAEGLANGPDVLETDGLQEVPLCSCRMETPKSREITTLANNQC
MATESVDHELGRCTNSVVKYELMRPSNKAPLLVLCEDHRGRMVKHQCCPGCGYFCTAGNF
MECQPESSISHRFHKDCASRVNNASYCPHCGEESSKAKEVTIAKADTTSTVTPVPGQEKG
SALEGRADTTTGSAAGPPLSEDDKLQGAASHVPEGFDPTGPAGLGRPTPGLSQGPGKETL
ESALIALDSEKPKKLRFHPKQLYFSARQGELQKVLLMLVDGIDPNFKMEHQNKRSPLHAA
AEAGHVDICHMLVQAGANIDTCSEDQRTPLMEAAENNHLEAVKYLIKAGALVDPKDAEGS
TCLHLAAKKGHYEVVQYLLSNGQMDVNCQDDGGWTPMIWATEYKHVDLVKLLLSKGSDIN
IRDNEENICLHWAAFSGCVDIAEILLAAKCDLHAVNIHGDSPLHIAARENRYDCVVLFLS
RDSDVTLKNKEGETPLQCASLNSQVWSALQMSKALQDSAPDRPSPVERIVSRDIARGYER
IPIPCVNAVDSEPCPSNYKYVSQNCVTSPMNIDRNITHLQYCVCIDDCSSSNCMCGQLSM
RCWYDKDGRLLPEFNMAEPPLIFECNHACSCWRNCRNRVVQNGLRARLQLYRTRDMGWGV
RSLQDIPPGTFVCEYVGELISDSEADVREEDSYLFDLDNKDGEVYCIDARFYGNVSRFIN
HHCEPNLVPVRVFMAHQDLRFPRIAFFSTRLIEAGEQLGFDYGERFWDIKGKLFSCRCGS
PKCRHSSAALAQRQASAAQEAQEDGLPDTSSAAAADPL
|
|
|
|---|
| BDBM50300032 |
|---|
| n/a |
|---|
| Name | BDBM50300032 |
|---|
| Synonyms: | 6,7-Dimethoxy-N-(1-methylpiperidin-4-yl)-2-morpholinoquinazolin-4-amine | CHEMBL571717 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H29N5O3 |
|---|
| Mol. Mass. | 387.476 |
|---|
| SMILES | COc1cc2nc(nc(NC3CCN(C)CC3)c2cc1OC)N1CCOCC1 |
|---|
| Structure | 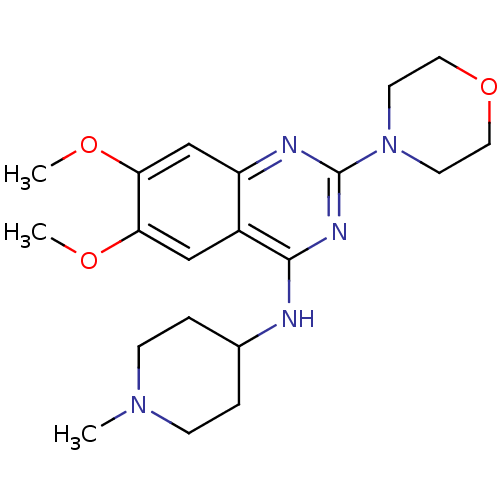
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Xiong, Y; Li, F; Babault, N; Dong, A; Zeng, H; Wu, H; Chen, X; Arrowsmith, CH; Brown, PJ; Liu, J; Vedadi, M; Jin, J Discovery of Potent and Selective Inhibitors for G9a-Like Protein (GLP) Lysine Methyltransferase. J Med Chem60:1876-1891 (2017) [PubMed] Article
Xiong, Y; Li, F; Babault, N; Dong, A; Zeng, H; Wu, H; Chen, X; Arrowsmith, CH; Brown, PJ; Liu, J; Vedadi, M; Jin, J Discovery of Potent and Selective Inhibitors for G9a-Like Protein (GLP) Lysine Methyltransferase. J Med Chem60:1876-1891 (2017) [PubMed] Article