| Reaction Details |
|---|
 | Report a problem with these data |
| Target | UDP-3-O-acyl-N-acetylglucosamine deacetylase |
|---|
| Ligand | BDBM50501752 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1699648 (CHEMBL4050630) |
|---|
| IC50 | 2.0±n/a nM |
|---|
| Citation |  Zhang, J; Chan, A; Lippa, B; Cross, JB; Liu, C; Yin, N; Romero, JA; Lawrence, J; Heney, R; Herradura, P; Goss, J; Clark, C; Abel, C; Zhang, Y; Poutsiaka, KM; Epie, F; Conrad, M; Mahamoon, A; Nguyen, K; Chavan, A; Clark, E; Li, TC; Cheng, RK; Wood, M; Andersen, OA; Brooks, M; Kwong, J; Barker, J; Parr, IB; Gu, Y; Ryan, MD; Coleman, S; Metcalf, CA Structure-based discovery of LpxC inhibitors. Bioorg Med Chem Lett27:1670-1680 (2017) [PubMed] Article Zhang, J; Chan, A; Lippa, B; Cross, JB; Liu, C; Yin, N; Romero, JA; Lawrence, J; Heney, R; Herradura, P; Goss, J; Clark, C; Abel, C; Zhang, Y; Poutsiaka, KM; Epie, F; Conrad, M; Mahamoon, A; Nguyen, K; Chavan, A; Clark, E; Li, TC; Cheng, RK; Wood, M; Andersen, OA; Brooks, M; Kwong, J; Barker, J; Parr, IB; Gu, Y; Ryan, MD; Coleman, S; Metcalf, CA Structure-based discovery of LpxC inhibitors. Bioorg Med Chem Lett27:1670-1680 (2017) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| UDP-3-O-acyl-N-acetylglucosamine deacetylase |
|---|
| Name: | UDP-3-O-acyl-N-acetylglucosamine deacetylase |
|---|
| Synonyms: | LPXC_ECOLI | UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase (LpxC) | UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase (LxpC) | UDP-3-O-acyl-GlcNAc deacetylase | asmB | envA | lpxC |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 33952.00 |
|---|
| Organism: | Escherichia coli |
|---|
| Description: | P0A725 |
|---|
| Residue: | 305 |
|---|
| Sequence: | MIKQRTLKRIVQATGVGLHTGKKVTLTLRPAPANTGVIYRRTDLNPPVDFPADAKSVRDT
MLCTCLVNEHDVRISTVEHLNAALAGLGIDNIVIEVNAPEIPIMDGSAAPFVYLLLDAGI
DELNCAKKFVRIKETVRVEDGDKWAEFKPYNGFSLDFTIDFNHPAIDSSNQRYAMNFSAD
AFMRQISRARTFGFMRDIEYLQSRGLCLGGSFDCAIVVDDYRVLNEDGLRFEDEFVRHKM
LDAIGDLFMCGHNIIGAFTAYKSGHALNNKLLQAVLAKQEAWEYVTFQDDAELPLAFKAP
SAVLA
|
|
|
|---|
| BDBM50501752 |
|---|
| n/a |
|---|
| Name | BDBM50501752 |
|---|
| Synonyms: | CHEMBL4095059 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C26H31N3O4 |
|---|
| Mol. Mass. | 449.542 |
|---|
| SMILES | C[C@@H](O)[C@H](NC1CCc2cc(ccc12)C#Cc1ccc(CN2CCOCC2)cc1)C(=O)NO |r| |
|---|
| Structure | 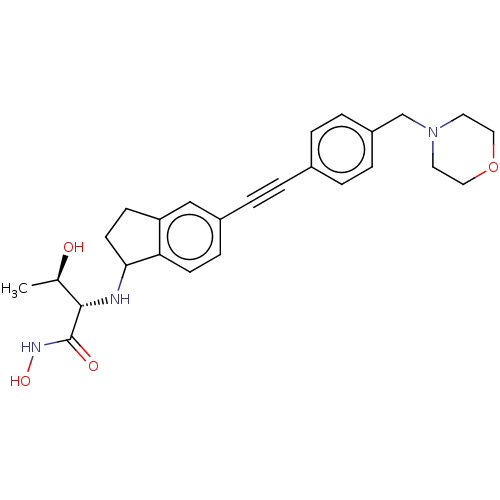
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Zhang, J; Chan, A; Lippa, B; Cross, JB; Liu, C; Yin, N; Romero, JA; Lawrence, J; Heney, R; Herradura, P; Goss, J; Clark, C; Abel, C; Zhang, Y; Poutsiaka, KM; Epie, F; Conrad, M; Mahamoon, A; Nguyen, K; Chavan, A; Clark, E; Li, TC; Cheng, RK; Wood, M; Andersen, OA; Brooks, M; Kwong, J; Barker, J; Parr, IB; Gu, Y; Ryan, MD; Coleman, S; Metcalf, CA Structure-based discovery of LpxC inhibitors. Bioorg Med Chem Lett27:1670-1680 (2017) [PubMed] Article
Zhang, J; Chan, A; Lippa, B; Cross, JB; Liu, C; Yin, N; Romero, JA; Lawrence, J; Heney, R; Herradura, P; Goss, J; Clark, C; Abel, C; Zhang, Y; Poutsiaka, KM; Epie, F; Conrad, M; Mahamoon, A; Nguyen, K; Chavan, A; Clark, E; Li, TC; Cheng, RK; Wood, M; Andersen, OA; Brooks, M; Kwong, J; Barker, J; Parr, IB; Gu, Y; Ryan, MD; Coleman, S; Metcalf, CA Structure-based discovery of LpxC inhibitors. Bioorg Med Chem Lett27:1670-1680 (2017) [PubMed] Article