

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Mitogen-activated protein kinase 9 | ||
| Ligand | BDBM50512318 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1848562 (CHEMBL4349103) | ||
| IC50 | 0.059000±n/a nM | ||
| Citation |  Riggs, JR; Elsner, J; Cashion, D; Robinson, D; Tehrani, L; Nagy, M; Fultz, KE; Krishna Narla, R; Peng, X; Tran, T; Kulkarni, A; Bahmanyar, S; Condroski, K; Pagarigan, B; Fenalti, G; LeBrun, L; Leftheris, K; Zhu, D; Boylan, JF Design and Optimization Leading to an Orally Active TTK Protein Kinase Inhibitor with Robust Single Agent Efficacy. J Med Chem62:4401-4410 (2019) [PubMed] Article Riggs, JR; Elsner, J; Cashion, D; Robinson, D; Tehrani, L; Nagy, M; Fultz, KE; Krishna Narla, R; Peng, X; Tran, T; Kulkarni, A; Bahmanyar, S; Condroski, K; Pagarigan, B; Fenalti, G; LeBrun, L; Leftheris, K; Zhu, D; Boylan, JF Design and Optimization Leading to an Orally Active TTK Protein Kinase Inhibitor with Robust Single Agent Efficacy. J Med Chem62:4401-4410 (2019) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Mitogen-activated protein kinase 9 | |||
| Name: | Mitogen-activated protein kinase 9 | ||
| Synonyms: | JNK-55 | JNK2 | JNK2/JNK3 | MAPK9 | MK09_HUMAN | Mitogen-Activated Protein Kinase 9 (JNK2) | Mitogen-activated protein kinase 8/9 | PRKM9 | SAPK1A | Stress-activated protein kinase JNK2 | c-Jun N-terminal kinase 2 | c-Jun N-terminal kinase 2 (JNK2) | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 48131.49 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | JNK-2 was purchased from Upstate Cell Signaling Solutions (formerly Upstate Biotechnology). | ||
| Residue: | 424 | ||
| Sequence: |
| ||
| BDBM50512318 | |||
| n/a | |||
| Name | BDBM50512318 | ||
| Synonyms: | CHEMBL4546504 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C25H29ClF3N5O4 | ||
| Mol. Mass. | 555.977 | ||
| SMILES | C[C@@H](Oc1cc(ccc1Nc1nc(O[C@H]2CC[C@@](C)(O)CC2)c2c(Cl)c[nH]c2n1)C(=O)N(C)C)C(F)(F)F |r,wU:17.19,14.14,1.0,(59.2,-50.35,;60.54,-51.12,;61.87,-50.34,;61.86,-48.8,;60.53,-48.04,;60.52,-46.49,;61.86,-45.72,;63.19,-46.49,;63.18,-48.03,;64.52,-48.8,;65.85,-48.03,;65.85,-46.49,;67.18,-45.72,;67.18,-44.18,;65.84,-43.41,;64.51,-44.19,;63.18,-43.42,;63.17,-41.87,;62.4,-40.53,;61.63,-41.87,;64.51,-41.11,;65.84,-41.87,;68.52,-46.48,;69.98,-46,;70.45,-44.53,;70.89,-47.24,;70,-48.49,;68.53,-48.02,;67.18,-48.8,;59.19,-45.72,;59.19,-44.18,;57.86,-46.49,;56.52,-45.72,;57.86,-48.03,;60.54,-52.66,;59.21,-53.43,;61.88,-53.42,;60.53,-54.19,)| | ||
| Structure | 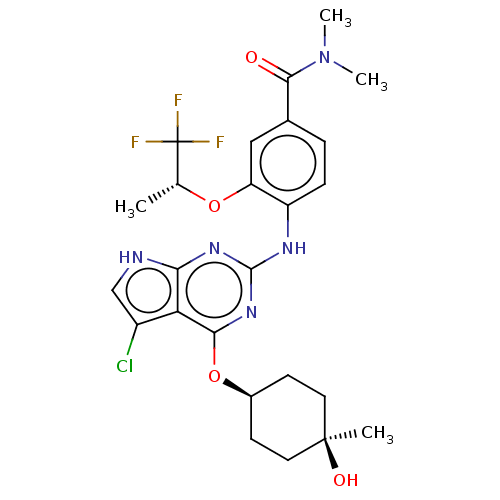
| ||