| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Cytochrome P450 3A4 |
|---|
| Ligand | BDBM268079 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1979602 (CHEMBL4612737) |
|---|
| IC50 | 7000±n/a nM |
|---|
| Citation |  Lücking, U; Wortmann, L; Wengner, AM; Lefranc, J; Lienau, P; Briem, H; Siemeister, G; Bömer, U; Denner, K; Schäfer, M; Koppitz, M; Eis, K; Bartels, F; Bader, B; Bone, W; Moosmayer, D; Holton, SJ; Eberspächer, U; Grudzinska-Goebel, J; Schatz, C; Deeg, G; Mumberg, D; von Nussbaum, F Damage Incorporated: Discovery of the Potent, Highly Selective, Orally Available ATR Inhibitor BAY 1895344 with Favorable Pharmacokinetic Properties and Promising Efficacy in Monotherapy and in Combination Treatments in Preclinical Tumor Models. J Med Chem63:7293-7325 (2020) [PubMed] Article Lücking, U; Wortmann, L; Wengner, AM; Lefranc, J; Lienau, P; Briem, H; Siemeister, G; Bömer, U; Denner, K; Schäfer, M; Koppitz, M; Eis, K; Bartels, F; Bader, B; Bone, W; Moosmayer, D; Holton, SJ; Eberspächer, U; Grudzinska-Goebel, J; Schatz, C; Deeg, G; Mumberg, D; von Nussbaum, F Damage Incorporated: Discovery of the Potent, Highly Selective, Orally Available ATR Inhibitor BAY 1895344 with Favorable Pharmacokinetic Properties and Promising Efficacy in Monotherapy and in Combination Treatments in Preclinical Tumor Models. J Med Chem63:7293-7325 (2020) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Cytochrome P450 3A4 |
|---|
| Name: | Cytochrome P450 3A4 |
|---|
| Synonyms: | Albendazole monooxygenase | Albendazole sulfoxidase | CP3A4_HUMAN | CYP3A3 | CYP3A4 | CYPIIIA3 | CYPIIIA4 | Cytochrome P450 3A3 | Cytochrome P450 3A4 (CYP3A4) | Cytochrome P450 HLp | Nifedipine oxidase | Quinine 3-monooxygenase | Taurochenodeoxycholate 6-alpha-hydroxylase |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 57349.57 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 503 |
|---|
| Sequence: | MALIPDLAMETWLLLAVSLVLLYLYGTHSHGLFKKLGIPGPTPLPFLGNILSYHKGFCMF
DMECHKKYGKVWGFYDGQQPVLAITDPDMIKTVLVKECYSVFTNRRPFGPVGFMKSAISI
AEDEEWKRLRSLLSPTFTSGKLKEMVPIIAQYGDVLVRNLRREAETGKPVTLKDVFGAYS
MDVITSTSFGVNIDSLNNPQDPFVENTKKLLRFDFLDPFFLSITVFPFLIPILEVLNICV
FPREVTNFLRKSVKRMKESRLEDTQKHRVDFLQLMIDSQNSKETESHKALSDLELVAQSI
IFIFAGYETTSSVLSFIMYELATHPDVQQKLQEEIDAVLPNKAPPTYDTVLQMEYLDMVV
NETLRLFPIAMRLERVCKKDVEINGMFIPKGVVVMIPSYALHRDPKYWTEPEKFLPERFS
KKNKDNIDPYIYTPFGSGPRNCIGMRFALMNMKLALIRVLQNFSFKPCKETQIPLKLSLG
GLLQPEKPVVLKVESRDGTVSGA
|
|
|
|---|
| BDBM268079 |
|---|
| n/a |
|---|
| Name | BDBM268079 |
|---|
| Synonyms: | 2-[(3R)-3-methylmorpholin-4-yl]-4-(1-methyl-1H-pyrazol-5-yl)-8-(1H-pyrazol-5-yl)-1,7-naphthyridine | US10772893, Example 111 | US11529356, Example 111 | US9549932, 111 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H21N7O |
|---|
| Mol. Mass. | 375.427 |
|---|
| SMILES | C[C@@H]1COCCN1c1cc(-c2ccnn2C)c2ccnc(-c3ccn[nH]3)c2n1 |r| |
|---|
| Structure | 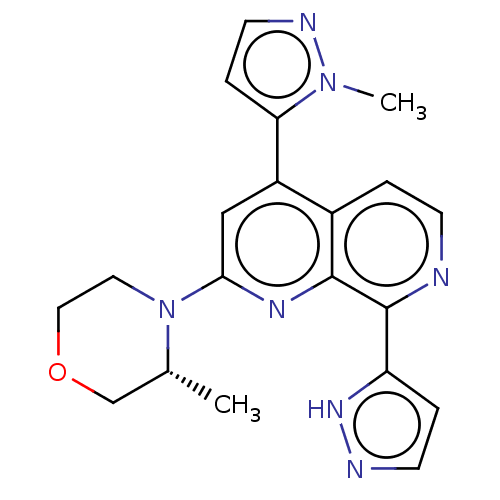
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Lücking, U; Wortmann, L; Wengner, AM; Lefranc, J; Lienau, P; Briem, H; Siemeister, G; Bömer, U; Denner, K; Schäfer, M; Koppitz, M; Eis, K; Bartels, F; Bader, B; Bone, W; Moosmayer, D; Holton, SJ; Eberspächer, U; Grudzinska-Goebel, J; Schatz, C; Deeg, G; Mumberg, D; von Nussbaum, F Damage Incorporated: Discovery of the Potent, Highly Selective, Orally Available ATR Inhibitor BAY 1895344 with Favorable Pharmacokinetic Properties and Promising Efficacy in Monotherapy and in Combination Treatments in Preclinical Tumor Models. J Med Chem63:7293-7325 (2020) [PubMed] Article
Lücking, U; Wortmann, L; Wengner, AM; Lefranc, J; Lienau, P; Briem, H; Siemeister, G; Bömer, U; Denner, K; Schäfer, M; Koppitz, M; Eis, K; Bartels, F; Bader, B; Bone, W; Moosmayer, D; Holton, SJ; Eberspächer, U; Grudzinska-Goebel, J; Schatz, C; Deeg, G; Mumberg, D; von Nussbaum, F Damage Incorporated: Discovery of the Potent, Highly Selective, Orally Available ATR Inhibitor BAY 1895344 with Favorable Pharmacokinetic Properties and Promising Efficacy in Monotherapy and in Combination Treatments in Preclinical Tumor Models. J Med Chem63:7293-7325 (2020) [PubMed] Article