| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Activin receptor type-1 |
|---|
| Ligand | BDBM50544399 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1992434 (CHEMBL4626169) |
|---|
| IC50 | 3.0±n/a nM |
|---|
| Citation |  Smil, D; Wong, JF; Williams, EP; Adamson, RJ; Howarth, A; McLeod, DA; Mamai, A; Kim, S; Wilson, BJ; Kiyota, T; Aman, A; Owen, J; Poda, G; Horiuchi, KY; Kuznetsova, E; Ma, H; Hamblin, JN; Cramp, S; Roberts, OG; Edwards, AM; Uehling, D; Al-Awar, R; Bullock, AN; O'Meara, JA; Isaac, MB Leveraging an Open Science Drug Discovery Model to Develop CNS-Penetrant ALK2 Inhibitors for the Treatment of Diffuse Intrinsic Pontine Glioma. J Med Chem63:10061-10085 (2020) [PubMed] Article Smil, D; Wong, JF; Williams, EP; Adamson, RJ; Howarth, A; McLeod, DA; Mamai, A; Kim, S; Wilson, BJ; Kiyota, T; Aman, A; Owen, J; Poda, G; Horiuchi, KY; Kuznetsova, E; Ma, H; Hamblin, JN; Cramp, S; Roberts, OG; Edwards, AM; Uehling, D; Al-Awar, R; Bullock, AN; O'Meara, JA; Isaac, MB Leveraging an Open Science Drug Discovery Model to Develop CNS-Penetrant ALK2 Inhibitors for the Treatment of Diffuse Intrinsic Pontine Glioma. J Med Chem63:10061-10085 (2020) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Activin receptor type-1 |
|---|
| Name: | Activin receptor type-1 |
|---|
| Synonyms: | 2.7.11.30 | ACTR-I | ACVR1 | ACVR1_HUMAN | ACVRLK2 | ALK-2 | ALK2/ACVR1 | Activin receptor type I | Activin receptor-like kinase 2 | Activin receptor-like kinase 2 (ALK-2) | Activin receptor-like kinase 2 (ALK2/ACVR1) | Q04771 | SKR1 | Serine/threonine-protein kinase receptor R1 | TGF-B superfamily receptor type I | TSR-I |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | 57158.32 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 509 |
|---|
| Sequence: | MVDGVMILPVLIMIALPSPSMEDEKPKVNPKLYMCVCEGLSCGNEDHCEGQQCFSSLSIN
DGFHVYQKGCFQVYEQGKMTCKTPPSPGQAVECCQGDWCNRNITAQLPTKGKSFPGTQNF
HLEVGLIILSVVFAVCLLACLLGVALRKFKRRNQERLNPRDVEYGTIEGLITTNVGDSTL
ADLLDHSCTSGSGSGLPFLVQRTVARQITLLECVGKGRYGEVWRGSWQGENVAVKIFSSR
DEKSWFRETELYNTVMLRHENILGFIASDMTSRHSSTQLWLITHYHEMGSLYDYLQLTTL
DTVSCLRIVLSIASGLAHLHIEIFGTQGKPAIAHRDLKSKNILVKKNGQCCIADLGLAVM
HSQSTNQLDVGNNPRVGTKRYMAPEVLDETIQVDCFDSYKRVDIWAFGLVLWEVARRMVS
NGIVEDYKPPFYDVVPNDPSFEDMRKVVCVDQQRPNIPNRWFSDPTLTSLAKLMKECWYQ
NPSARLTALRIKKTLTKIDNSLDKLKTDC
|
|
|
|---|
| BDBM50544399 |
|---|
| n/a |
|---|
| Name | BDBM50544399 |
|---|
| Synonyms: | CHEMBL4638273 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C25H26N4O3 |
|---|
| Mol. Mass. | 430.4989 |
|---|
| SMILES | COc1cc(cc(OC)c1OC)-c1cncc(-c2ccc(cc2)N2CCNCC2)c1C#N |
|---|
| Structure | 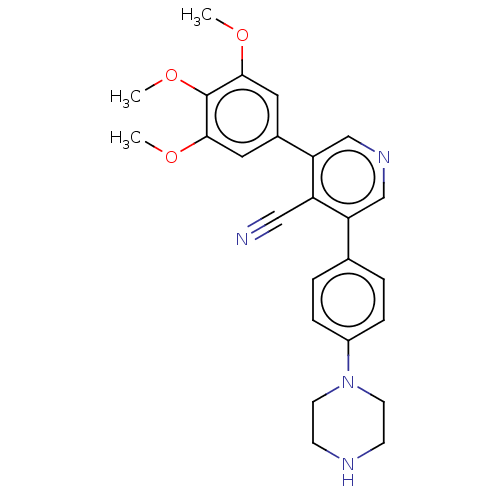
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Smil, D; Wong, JF; Williams, EP; Adamson, RJ; Howarth, A; McLeod, DA; Mamai, A; Kim, S; Wilson, BJ; Kiyota, T; Aman, A; Owen, J; Poda, G; Horiuchi, KY; Kuznetsova, E; Ma, H; Hamblin, JN; Cramp, S; Roberts, OG; Edwards, AM; Uehling, D; Al-Awar, R; Bullock, AN; O'Meara, JA; Isaac, MB Leveraging an Open Science Drug Discovery Model to Develop CNS-Penetrant ALK2 Inhibitors for the Treatment of Diffuse Intrinsic Pontine Glioma. J Med Chem63:10061-10085 (2020) [PubMed] Article
Smil, D; Wong, JF; Williams, EP; Adamson, RJ; Howarth, A; McLeod, DA; Mamai, A; Kim, S; Wilson, BJ; Kiyota, T; Aman, A; Owen, J; Poda, G; Horiuchi, KY; Kuznetsova, E; Ma, H; Hamblin, JN; Cramp, S; Roberts, OG; Edwards, AM; Uehling, D; Al-Awar, R; Bullock, AN; O'Meara, JA; Isaac, MB Leveraging an Open Science Drug Discovery Model to Develop CNS-Penetrant ALK2 Inhibitors for the Treatment of Diffuse Intrinsic Pontine Glioma. J Med Chem63:10061-10085 (2020) [PubMed] Article