

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Mitogen-activated protein kinase 9 | ||
| Ligand | BDBM50105742 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEBML_91768 | ||
| IC50 | 5±n/a nM | ||
| Citation |  Adams, JL; Boehm, JC; Gallagher, TF; Kassis, S; Webb, EF; Hall, R; Sorenson, M; Garigipati, R; Griswold, DE; Lee, JC Pyrimidinylimidazole inhibitors of p38: cyclic N-1 imidazole substituents enhance p38 kinase inhibition and oral activity. Bioorg Med Chem Lett11:2867-70 (2001) [PubMed] Adams, JL; Boehm, JC; Gallagher, TF; Kassis, S; Webb, EF; Hall, R; Sorenson, M; Garigipati, R; Griswold, DE; Lee, JC Pyrimidinylimidazole inhibitors of p38: cyclic N-1 imidazole substituents enhance p38 kinase inhibition and oral activity. Bioorg Med Chem Lett11:2867-70 (2001) [PubMed] | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Mitogen-activated protein kinase 9 | |||
| Name: | Mitogen-activated protein kinase 9 | ||
| Synonyms: | JNK-55 | JNK2 | JNK2/JNK3 | MAPK9 | MK09_HUMAN | Mitogen-Activated Protein Kinase 9 (JNK2) | Mitogen-activated protein kinase 8/9 | PRKM9 | SAPK1A | Stress-activated protein kinase JNK2 | c-Jun N-terminal kinase 2 | c-Jun N-terminal kinase 2 (JNK2) | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 48131.49 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | JNK-2 was purchased from Upstate Cell Signaling Solutions (formerly Upstate Biotechnology). | ||
| Residue: | 424 | ||
| Sequence: |
| ||
| BDBM50105742 | |||
| n/a | |||
| Name | BDBM50105742 | ||
| Synonyms: | 4-[4-(4-Fluoro-phenyl)-5-(2-methoxy-pyrimidin-4-yl)-imidazol-1-yl]-cyclohexanol | CHEMBL97162 | trans -1-(4-hydroxycyclohexyl)-4-(4-fluorophenyl)-5-(2-methoxypyrimidin-4-yl)imidazole | ||
| Type | Small organic molecule | ||
| Emp. Form. | C20H21FN4O2 | ||
| Mol. Mass. | 368.4047 | ||
| SMILES | COc1nccc(n1)-c1c(ncn1[C@H]1CC[C@H](O)CC1)-c1ccc(F)cc1 |wU:16.18,wD:13.14,(3.13,-3.72,;3.16,-2.18,;4.5,-1.42,;4.53,.13,;5.87,.86,;7.2,.06,;7.17,-1.48,;5.82,-2.22,;8.5,-2.27,;8.7,-3.8,;10.21,-4.09,;10.93,-2.74,;9.88,-1.62,;10.17,-.1,;11.64,.4,;11.93,1.91,;10.77,2.91,;11.06,4.43,;9.31,2.4,;9.02,.89,;7.63,-4.91,;8.06,-6.39,;6.98,-7.48,;5.49,-7.1,;4.43,-8.22,;5.06,-5.63,;6.14,-4.53,)| | ||
| Structure | 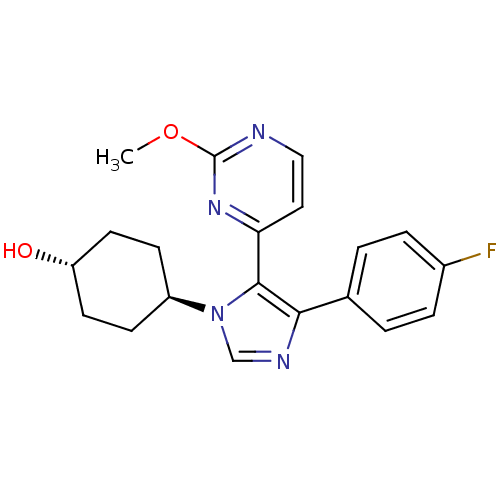
| ||