

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Myc proto-oncogene protein | ||
| Ligand | BDBM50579852 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_2145528 (CHEMBL5029808) | ||
| IC50 | 2.4±n/a nM | ||
| Citation |  Hill, MD; Quesnelle, C; Tokarski, J; Fang, H; Fanslau, C; Haarhoff, Z; Kramer, M; Madari, S; Wiebesiek, A; Morrison, J; Simmermacher-Mayer, J; Sinz, M; Westhouse, R; Xie, C; Zhao, J; Huang, L; Sheriff, S; Yan, C; Marsilio, F; Everlof, G; Zvyaga, T; Lee, F; Gavai, AV; Degnan, AP Development of BET inhibitors as potential treatments for cancer: A new carboline chemotype. Bioorg Med Chem Lett51:0 (2021) [PubMed] Article Hill, MD; Quesnelle, C; Tokarski, J; Fang, H; Fanslau, C; Haarhoff, Z; Kramer, M; Madari, S; Wiebesiek, A; Morrison, J; Simmermacher-Mayer, J; Sinz, M; Westhouse, R; Xie, C; Zhao, J; Huang, L; Sheriff, S; Yan, C; Marsilio, F; Everlof, G; Zvyaga, T; Lee, F; Gavai, AV; Degnan, AP Development of BET inhibitors as potential treatments for cancer: A new carboline chemotype. Bioorg Med Chem Lett51:0 (2021) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Myc proto-oncogene protein | |||
| Name: | Myc proto-oncogene protein | ||
| Synonyms: | BHLHE39 | Class E basic helix-loop-helix protein 39 | MYC | MYC_HUMAN | Proto-oncogene c-Myc | Transcription factor p64 | ||
| Type: | PROTEIN | ||
| Mol. Mass.: | 48794.95 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | ChEMBL_935266 | ||
| Residue: | 439 | ||
| Sequence: |
| ||
| BDBM50579852 | |||
| n/a | |||
| Name | BDBM50579852 | ||
| Synonyms: | CHEMBL5090513 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C28H31FN6O3 | ||
| Mol. Mass. | 521.601 | ||
| SMILES | [2H]C([2H])([2H])c1nnn(C)c1-c1cc2n([C@@H](C3CCOCC3)c3cc(C)on3)c3cc(cnc3c2cc1F)C(C)(C)O |r,wD:14.22,(33.12,-33.94,;34.66,-33.94,;35.42,-35.27,;34.12,-35.06,;35.39,-32.58,;36.91,-32.38,;37.19,-30.86,;35.83,-30.13,;35.62,-28.61,;34.72,-31.2,;33.21,-30.93,;32.21,-32.1,;30.7,-31.82,;29.49,-32.77,;29.54,-34.31,;28.23,-35.12,;26.88,-34.39,;25.58,-35.2,;25.62,-36.74,;26.98,-37.47,;28.29,-36.66,;30.9,-35.03,;31.11,-36.55,;32.63,-36.82,;33.3,-38.2,;33.35,-35.46,;32.29,-34.35,;28.21,-31.9,;26.73,-32.26,;25.68,-31.16,;26.11,-29.69,;27.6,-29.33,;28.65,-30.44,;30.18,-30.37,;31.17,-29.2,;32.68,-29.47,;33.67,-28.29,;24.18,-31.53,;23.75,-33.01,;22.68,-31.92,;23.12,-30.42,)| | ||
| Structure | 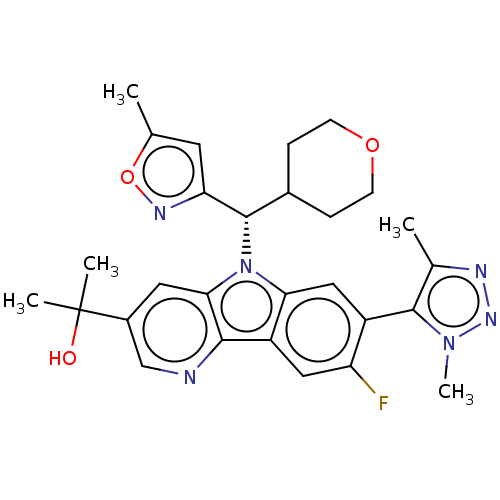
| ||