

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Isocitrate dehydrogenase [NADP] cytoplasmic | ||
| Ligand | BDBM416155 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_2165763 (CHEMBL5050624) | ||
| IC50 | 16±n/a nM | ||
| Citation |  Huang, C; Fischer, C; Machacek, MR; Bogen, S; Biftu, T; Huang, X; Reutershan, MH; Otte, R; Hong, Q; Wu, Z; Yu, Y; Park, M; Chen, L; Biju, P; Knemeyer, I; Lu, P; Kochansky, CJ; Hicks, MB; Liu, Y; Helmy, R; Fradera, X; Donofrio, A; Close, J; Maddess, ML; White, C; Sloman, DL; Sciammetta, N; Lu, J; Gibeau, C; Simov, V; Zhang, H; Fuller, P; Witter, D Diminishing GSH-Adduct Formation of Tricyclic Diazepine-based Mutant IDH1 Inhibitors. ACS Med Chem Lett13:734-741 (2022) [PubMed] Article Huang, C; Fischer, C; Machacek, MR; Bogen, S; Biftu, T; Huang, X; Reutershan, MH; Otte, R; Hong, Q; Wu, Z; Yu, Y; Park, M; Chen, L; Biju, P; Knemeyer, I; Lu, P; Kochansky, CJ; Hicks, MB; Liu, Y; Helmy, R; Fradera, X; Donofrio, A; Close, J; Maddess, ML; White, C; Sloman, DL; Sciammetta, N; Lu, J; Gibeau, C; Simov, V; Zhang, H; Fuller, P; Witter, D Diminishing GSH-Adduct Formation of Tricyclic Diazepine-based Mutant IDH1 Inhibitors. ACS Med Chem Lett13:734-741 (2022) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Isocitrate dehydrogenase [NADP] cytoplasmic | |||
| Name: | Isocitrate dehydrogenase [NADP] cytoplasmic | ||
| Synonyms: | Cytosolic NADP-isocitrate dehydrogenase | IDH | IDH1 | IDHC_HUMAN | IDP | Isocitrate Dehydrogenase (IDH1) | Isocitrate dehydrogenase 1 (IDH1) | NADP(+)-specific ICDH | Oxalosuccinate decarboxylase | PICD | ||
| Type: | Protein | ||
| Mol. Mass.: | 46661.29 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | O75874 | ||
| Residue: | 414 | ||
| Sequence: |
| ||
| BDBM416155 | |||
| n/a | |||
| Name | BDBM416155 | ||
| Synonyms: | 6-{[trans-4-(1- methylethoxy)cyclohexyl]car- bonyl}-8-[(1S,4S)-2-oxa-5- azabicyclo[2.2.1]hept-5-yl]- 6,11-dihydro-5H-pyrido[2,3- b][1,5]benzodiazepine | US10442819, Example 300 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C27H34N4O3 | ||
| Mol. Mass. | 462.5839 | ||
| SMILES | CC(C)O[C@H]1CC[C@@H](CC1)C(=O)N1Cc2cccnc2Nc2ccc(cc12)N1C[C@@H]2C[C@H]1CO2 |r,wU:7.10,29.33,wD:4.3,31.34,THB:24:27:32.33:30,(6.2,5.88,;5.43,4.54,;6.2,3.21,;3.89,4.54,;3.12,3.21,;1.58,3.21,;.81,1.88,;1.58,.54,;3.12,.54,;3.89,1.88,;.81,-.79,;-.73,-.79,;1.58,-2.12,;3.12,-2.12,;4.08,-3.33,;5.55,-2.87,;6.68,-3.92,;6.34,-5.42,;4.87,-5.88,;3.74,-4.83,;2.35,-5.5,;.97,-4.83,;-.16,-5.88,;-1.64,-5.42,;-1.98,-3.92,;-.85,-2.87,;.62,-3.33,;-3.47,-3.52,;-3.86,-2.04,;-5.2,-2.8,;-6.68,-2.41,;-4.8,-4.29,;-5.57,-5.63,;-5.96,-4.14,)| | ||
| Structure | 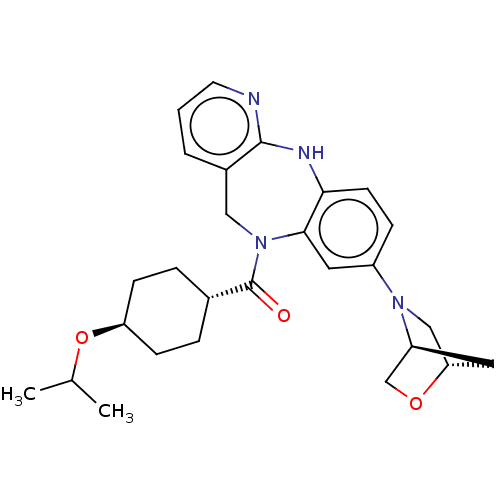
| ||