| Citation |  Frankowski, KJ; Patnaik, S; Wang, C; Southall, N; Dutta, D; De, S; Li, D; Dextras, C; Lin, YH; Bryant-Connah, M; Davis, D; Wang, F; Wachsmuth, LM; Shah, P; Williams, J; Kabir, M; Zhu, E; Baljinnyam, B; Wang, A; Xu, X; Norton, J; Ferrer, M; Titus, S; Simeonov, A; Zheng, W; Mathews Griner, LA; Jadhav, A; Aubé, J; Henderson, MJ; Rudloff, U; Schoenen, FJ; Huang, S; Marugan, JJ Discovery and Optimization of Pyrrolopyrimidine Derivatives as Selective Disruptors of the Perinucleolar Compartment, a Marker of Tumor Progression toward Metastasis. J Med Chem65:8303-8331 (2022) [PubMed] Article Frankowski, KJ; Patnaik, S; Wang, C; Southall, N; Dutta, D; De, S; Li, D; Dextras, C; Lin, YH; Bryant-Connah, M; Davis, D; Wang, F; Wachsmuth, LM; Shah, P; Williams, J; Kabir, M; Zhu, E; Baljinnyam, B; Wang, A; Xu, X; Norton, J; Ferrer, M; Titus, S; Simeonov, A; Zheng, W; Mathews Griner, LA; Jadhav, A; Aubé, J; Henderson, MJ; Rudloff, U; Schoenen, FJ; Huang, S; Marugan, JJ Discovery and Optimization of Pyrrolopyrimidine Derivatives as Selective Disruptors of the Perinucleolar Compartment, a Marker of Tumor Progression toward Metastasis. J Med Chem65:8303-8331 (2022) [PubMed] Article |
|---|
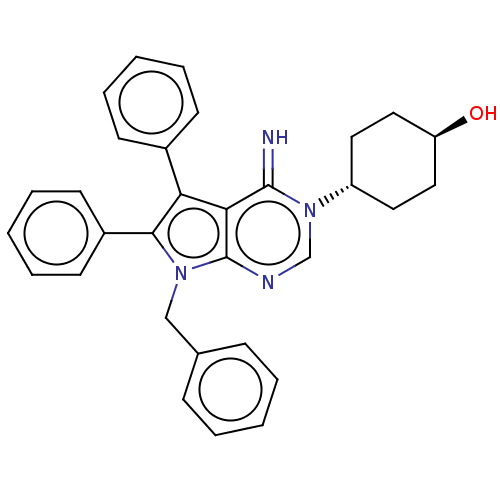
| SMILES | O[C@H]1CC[C@@H](CC1)n1cnc2n(Cc3ccccc3)c(c(-c3ccccc3)c2c1=N)-c1ccccc1 |wU:4.7,wD:1.0,(20.99,-11.86,;22.33,-11.09,;23.66,-11.86,;25,-11.09,;25,-9.55,;23.66,-8.78,;22.33,-9.55,;26.33,-8.78,;26.33,-7.24,;27.66,-6.47,;29,-7.24,;30.46,-6.76,;30.94,-5.3,;29.91,-4.15,;30.38,-2.69,;29.35,-1.55,;27.85,-1.87,;27.37,-3.33,;28.4,-4.47,;31.37,-8.01,;30.46,-9.26,;30.94,-10.72,;32.44,-11.04,;32.92,-12.5,;31.89,-13.65,;30.38,-13.33,;29.91,-11.86,;29,-8.78,;27.66,-9.55,;27.66,-11.09,;32.91,-8.01,;33.68,-6.68,;35.22,-6.68,;35.99,-8.01,;35.22,-9.34,;33.68,-9.34,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Frankowski, KJ; Patnaik, S; Wang, C; Southall, N; Dutta, D; De, S; Li, D; Dextras, C; Lin, YH; Bryant-Connah, M; Davis, D; Wang, F; Wachsmuth, LM; Shah, P; Williams, J; Kabir, M; Zhu, E; Baljinnyam, B; Wang, A; Xu, X; Norton, J; Ferrer, M; Titus, S; Simeonov, A; Zheng, W; Mathews Griner, LA; Jadhav, A; Aubé, J; Henderson, MJ; Rudloff, U; Schoenen, FJ; Huang, S; Marugan, JJ Discovery and Optimization of Pyrrolopyrimidine Derivatives as Selective Disruptors of the Perinucleolar Compartment, a Marker of Tumor Progression toward Metastasis. J Med Chem65:8303-8331 (2022) [PubMed] Article
Frankowski, KJ; Patnaik, S; Wang, C; Southall, N; Dutta, D; De, S; Li, D; Dextras, C; Lin, YH; Bryant-Connah, M; Davis, D; Wang, F; Wachsmuth, LM; Shah, P; Williams, J; Kabir, M; Zhu, E; Baljinnyam, B; Wang, A; Xu, X; Norton, J; Ferrer, M; Titus, S; Simeonov, A; Zheng, W; Mathews Griner, LA; Jadhav, A; Aubé, J; Henderson, MJ; Rudloff, U; Schoenen, FJ; Huang, S; Marugan, JJ Discovery and Optimization of Pyrrolopyrimidine Derivatives as Selective Disruptors of the Perinucleolar Compartment, a Marker of Tumor Progression toward Metastasis. J Med Chem65:8303-8331 (2022) [PubMed] Article