

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Alpha-1A adrenergic receptor | ||
| Ligand | BDBM50302568 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_594566 (CHEMBL1037061) | ||
| IC50 | 3200±n/a nM | ||
| Citation |  Semple, G; Tran, TA; Kramer, B; Hsu, D; Han, S; Choi, J; Vallar, P; Casper, MD; Zou, N; Hauser, EK; Thomsen, W; Whelan, K; Sengupta, D; Morgan, M; Sekiguchi, Y; Kanuma, K; Chaki, S; Grottick, AJ Pyrimidine-based antagonists of h-MCH-R1 derived from ATC0175: in vitro profiling and in vivo evaluation. Bioorg Med Chem Lett19:6166-71 (2009) [PubMed] Article Semple, G; Tran, TA; Kramer, B; Hsu, D; Han, S; Choi, J; Vallar, P; Casper, MD; Zou, N; Hauser, EK; Thomsen, W; Whelan, K; Sengupta, D; Morgan, M; Sekiguchi, Y; Kanuma, K; Chaki, S; Grottick, AJ Pyrimidine-based antagonists of h-MCH-R1 derived from ATC0175: in vitro profiling and in vivo evaluation. Bioorg Med Chem Lett19:6166-71 (2009) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Alpha-1A adrenergic receptor | |||
| Name: | Alpha-1A adrenergic receptor | ||
| Synonyms: | ADA1A_HUMAN | ADRA1A | ADRA1C | Adrenergic alpha1A | Alpha 1A-adrenoceptor | Alpha 1A-adrenoreceptor | Alpha adrenergic receptor 1a | Alpha-1C adrenergic receptor | Alpha-adrenergic receptor 1c | Cerebral cortex alpha adrenergic receptor | adrenergic Alpha1 | adrenergic Alpha1C | ||
| Type: | Cell-surface receptors | ||
| Mol. Mass.: | 51511.67 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P35348 | ||
| Residue: | 466 | ||
| Sequence: |
| ||
| BDBM50302568 | |||
| n/a | |||
| Name | BDBM50302568 | ||
| Synonyms: | CHEMBL566075 | N-[(cis-4-{[4-(Dimethylamino)-5-methylpyrimidin-2-yl]amino}cyclohexyl)methyl]-3,5-bis(trifluoromethyl)benzamide | ||
| Type | Small organic molecule | ||
| Emp. Form. | C23H27F6N5O | ||
| Mol. Mass. | 503.4838 | ||
| SMILES | CN(C)c1nc(N[C@@H]2CC[C@H](CNC(=O)c3cc(cc(c3)C(F)(F)F)C(F)(F)F)CC2)ncc1C |r,wU:10.10,7.6,(-4.66,-23.98,;-5.99,-24.76,;-7.33,-23.99,;-5.98,-26.3,;-4.65,-27.06,;-4.64,-28.61,;-3.31,-29.38,;-1.98,-28.61,;-.65,-29.38,;.68,-28.62,;.69,-27.08,;2.02,-26.31,;3.36,-27.08,;4.69,-26.32,;4.7,-24.78,;6.02,-27.09,;7.35,-26.32,;8.69,-27.09,;8.69,-28.63,;7.36,-29.4,;6.03,-28.62,;7.35,-30.94,;6.01,-31.7,;8.68,-31.72,;5.86,-30.53,;10.03,-26.32,;11.36,-27.09,;10.03,-24.78,;10.43,-27.81,;-.65,-26.31,;-1.99,-27.07,;-5.98,-29.38,;-7.32,-28.61,;-7.31,-27.07,;-8.65,-26.3,)| | ||
| Structure | 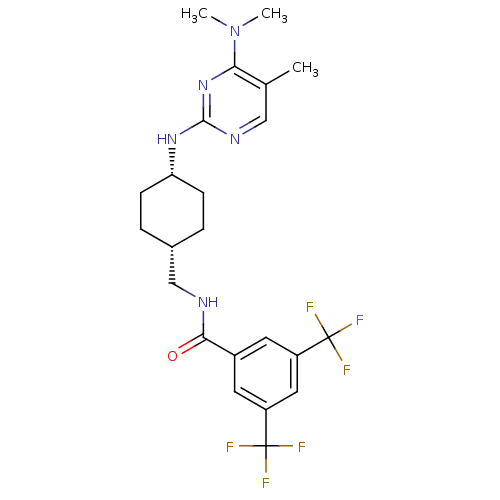
| ||