| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Peroxisome proliferator-activated receptor delta |
|---|
| Ligand | BDBM50312477 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_620660 (CHEMBL1115681) |
|---|
| EC50 | 30±n/a nM |
|---|
| Citation |  Epple, R; Cow, C; Xie, Y; Azimioara, M; Russo, R; Wang, X; Wityak, J; Karanewsky, DS; Tuntland, T; Nguyêñ-Trân, VT; Cuc Ngo, C; Huang, D; Saez, E; Spalding, T; Gerken, A; Iskandar, M; Seidel, HM; Tian, SS Novel bisaryl substituted thiazoles and oxazoles as highly potent and selective peroxisome proliferator-activated receptor delta agonists. J Med Chem53:77-105 (2010) [PubMed] Article Epple, R; Cow, C; Xie, Y; Azimioara, M; Russo, R; Wang, X; Wityak, J; Karanewsky, DS; Tuntland, T; Nguyêñ-Trân, VT; Cuc Ngo, C; Huang, D; Saez, E; Spalding, T; Gerken, A; Iskandar, M; Seidel, HM; Tian, SS Novel bisaryl substituted thiazoles and oxazoles as highly potent and selective peroxisome proliferator-activated receptor delta agonists. J Med Chem53:77-105 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Peroxisome proliferator-activated receptor delta |
|---|
| Name: | Peroxisome proliferator-activated receptor delta |
|---|
| Synonyms: | NUC1 | Nr1c2 | Nuclear hormone receptor 1 | Nuclear receptor subfamily 1 group C member 2 | PPAR-beta | PPAR-delta | PPARD_MOUSE | Pparb | Ppard |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 49719.14 |
|---|
| Organism: | Mus musculus |
|---|
| Description: | ChEMBL_1288931 |
|---|
| Residue: | 440 |
|---|
| Sequence: | MEQPQEETPEAREEEKEEVAMGDGAPELNGGPEHTLPSSSCADLSQNSSPSSLLDQLQMG
CDGASGGSLNMECRVCGDKASGFHYGVHACEGCKGFFRRTIRMKLEYEKCDRICKIQKKN
RNKCQYCRFQKCLALGMSHNAIRFGRMPEAEKRKLVAGLTASEGCQHNPQLADLKAFSKH
IYNAYLKNFNMTKKKARSILTGKSSHNAPFVIHDIETLWQAEKGLVWKQLVNGLPPYNEI
SVHVFYRCQSTTVETVRELTEFAKNIPNFSSLFLNDQVTLLKYGVHEAIFAMLASIVNKD
GLLVANGSGFVTHEFLRSLRKPFSDIIEPKFEFAVKFNALELDDSDLALFIAAIILCGDR
PGLMNVPQVEAIQDTILRALEFHLQVNHPDSQYLFPKLLQKMADLRQLVTEHAQMMQWLK
KTESETLLHPLLQEIYKDMY
|
|
|
|---|
| BDBM50312477 |
|---|
| n/a |
|---|
| Name | BDBM50312477 |
|---|
| Synonyms: | 2-(4-((4-(2-Methoxypyrimidin-5-yl)-5-(4-propylphenyl)oxazol-2-yl)methoxy)-2-methylphenoxy)acetic Acid | CHEMBL1080109 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C27H27N3O6 |
|---|
| Mol. Mass. | 489.5198 |
|---|
| SMILES | CCCc1ccc(cc1)-c1oc(COc2ccc(OCC(O)=O)c(C)c2)nc1-c1cnc(OC)nc1 |
|---|
| Structure | 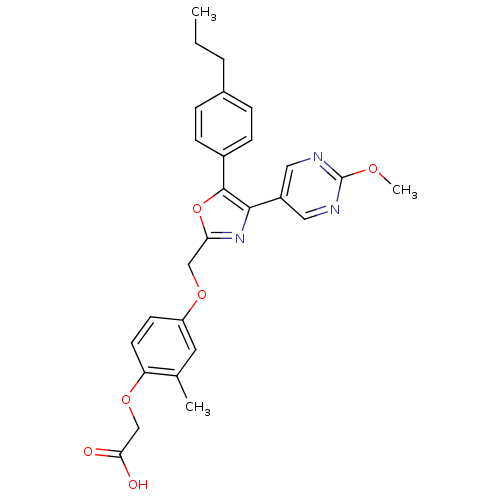
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Epple, R; Cow, C; Xie, Y; Azimioara, M; Russo, R; Wang, X; Wityak, J; Karanewsky, DS; Tuntland, T; Nguyêñ-Trân, VT; Cuc Ngo, C; Huang, D; Saez, E; Spalding, T; Gerken, A; Iskandar, M; Seidel, HM; Tian, SS Novel bisaryl substituted thiazoles and oxazoles as highly potent and selective peroxisome proliferator-activated receptor delta agonists. J Med Chem53:77-105 (2010) [PubMed] Article
Epple, R; Cow, C; Xie, Y; Azimioara, M; Russo, R; Wang, X; Wityak, J; Karanewsky, DS; Tuntland, T; Nguyêñ-Trân, VT; Cuc Ngo, C; Huang, D; Saez, E; Spalding, T; Gerken, A; Iskandar, M; Seidel, HM; Tian, SS Novel bisaryl substituted thiazoles and oxazoles as highly potent and selective peroxisome proliferator-activated receptor delta agonists. J Med Chem53:77-105 (2010) [PubMed] Article