| Reaction Details |
|---|
 | Report a problem with these data |
| Target | cAMP-specific 3',5'-cyclic phosphodiesterase 4B |
|---|
| Ligand | BDBM50328107 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_665324 (CHEMBL1261001) |
|---|
| IC50 | 40±n/a nM |
|---|
| Citation |  Taltavull, J; Serrat, J; Gràcia, J; Gavaldà, A; Andrés, M; Córdoba, M; Miralpeix, M; Vilella, D; Beleta, J; Ryder, H; Pagès, L Synthesis and biological activity of pyrido[3',2':4,5]thieno[3,2-d]pyrimidines as phosphodiesterase type 4 inhibitors. J Med Chem53:6912-22 (2010) [PubMed] Article Taltavull, J; Serrat, J; Gràcia, J; Gavaldà, A; Andrés, M; Córdoba, M; Miralpeix, M; Vilella, D; Beleta, J; Ryder, H; Pagès, L Synthesis and biological activity of pyrido[3',2':4,5]thieno[3,2-d]pyrimidines as phosphodiesterase type 4 inhibitors. J Med Chem53:6912-22 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| cAMP-specific 3',5'-cyclic phosphodiesterase 4B |
|---|
| Name: | cAMP-specific 3',5'-cyclic phosphodiesterase 4B |
|---|
| Synonyms: | 3',5'-cyclic phosphodiesterase | DPDE4 | Isoform PDE4B1 | PDE32 | PDE4B | PDE4B1 | PDE4B_HUMAN | Phosphodiesterase 4B | Phosphodiesterase 4B (PDE4B) | Phosphodiesterase 4B (PDE4B1) | Phosphodiesterase Type 4 (PDE4B) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 83318.87 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q07343 |
|---|
| Residue: | 736 |
|---|
| Sequence: | MKKSRSVMTVMADDNVKDYFECSLSKSYSSSSNTLGIDLWRGRRCCSGNLQLPPLSQRQS
ERARTPEGDGISRPTTLPLTTLPSIAITTVSQECFDVENGPSPGRSPLDPQASSSAGLVL
HATFPGHSQRRESFLYRSDSDYDLSPKAMSRNSSLPSEQHGDDLIVTPFAQVLASLRSVR
NNFTILTNLHGTSNKRSPAASQPPVSRVNPQEESYQKLAMETLEELDWCLDQLETIQTYR
SVSEMASNKFKRMLNRELTHLSEMSRSGNQVSEYISNTFLDKQNDVEIPSPTQKDREKKK
KQQLMTQISGVKKLMHSSSLNNTSISRFGVNTENEDHLAKELEDLNKWGLNIFNVAGYSH
NRPLTCIMYAIFQERDLLKTFRISSDTFITYMMTLEDHYHSDVAYHNSLHAADVAQSTHV
LLSTPALDAVFTDLEILAAIFAAAIHDVDHPGVSNQFLINTNSELALMYNDESVLENHHL
AVGFKLLQEEHCDIFMNLTKKQRQTLRKMVIDMVLATDMSKHMSLLADLKTMVETKKVTS
SGVLLLDNYTDRIQVLRNMVHCADLSNPTKSLELYRQWTDRIMEEFFQQGDKERERGMEI
SPMCDKHTASVEKSQVGFIDYIVHPLWETWADLVQPDAQDILDTLEDNRNWYQSMIPQSP
SPPLDEQNRDCQGLMEKFQFELTLDEEDSEGPEKEGEGHSYFSSTKTLCVIDPENRDSLG
ETDIDIATEDKSPVDT
|
|
|
|---|
| BDBM50328107 |
|---|
| n/a |
|---|
| Name | BDBM50328107 |
|---|
| Synonyms: | CHEMBL1257655 | N-(2-Methoxybenzyl)-2,2-dimethyl-5-morpholin-4-yl-1,4-dihydro-2H-pyrano[4'',3'':4',5']thieno[3,2-d]pyrimidin-8-amine |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C26H29N5O3S |
|---|
| Mol. Mass. | 491.605 |
|---|
| SMILES | COc1ccccc1CNc1ncnc2c1sc1nc(N3CCOCC3)c3COC(C)(C)Cc3c21 |
|---|
| Structure | 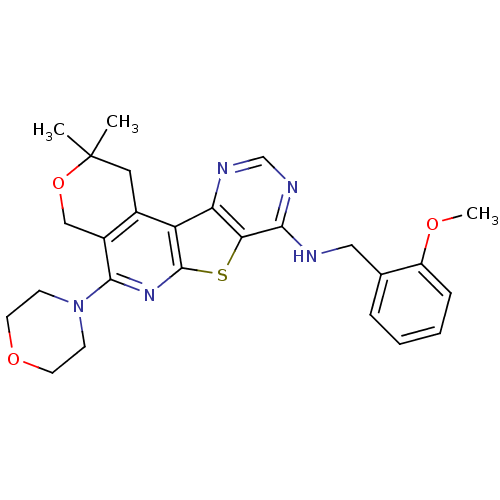
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Taltavull, J; Serrat, J; Gràcia, J; Gavaldà, A; Andrés, M; Córdoba, M; Miralpeix, M; Vilella, D; Beleta, J; Ryder, H; Pagès, L Synthesis and biological activity of pyrido[3',2':4,5]thieno[3,2-d]pyrimidines as phosphodiesterase type 4 inhibitors. J Med Chem53:6912-22 (2010) [PubMed] Article
Taltavull, J; Serrat, J; Gràcia, J; Gavaldà, A; Andrés, M; Córdoba, M; Miralpeix, M; Vilella, D; Beleta, J; Ryder, H; Pagès, L Synthesis and biological activity of pyrido[3',2':4,5]thieno[3,2-d]pyrimidines as phosphodiesterase type 4 inhibitors. J Med Chem53:6912-22 (2010) [PubMed] Article