| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Bifunctional epoxide hydrolase 2 |
|---|
| Ligand | BDBM50335966 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_716996 (CHEMBL1671068) |
|---|
| IC50 | 1.9±n/a nM |
|---|
| Citation |  Anandan, SK; Webb, HK; Chen, D; Wang, YX; Aavula, BR; Cases, S; Cheng, Y; Do, ZN; Mehra, U; Tran, V; Vincelette, J; Waszczuk, J; White, K; Wong, KR; Zhang, LN; Jones, PD; Hammock, BD; Patel, DV; Whitcomb, R; MacIntyre, DE; Sabry, J; Gless, R 1-(1-acetyl-piperidin-4-yl)-3-adamantan-1-yl-urea (AR9281) as a potent, selective, and orally available soluble epoxide hydrolase inhibitor with efficacy in rodent models of hypertension and dysglycemia. Bioorg Med Chem Lett21:983-8 (2011) [PubMed] Article Anandan, SK; Webb, HK; Chen, D; Wang, YX; Aavula, BR; Cases, S; Cheng, Y; Do, ZN; Mehra, U; Tran, V; Vincelette, J; Waszczuk, J; White, K; Wong, KR; Zhang, LN; Jones, PD; Hammock, BD; Patel, DV; Whitcomb, R; MacIntyre, DE; Sabry, J; Gless, R 1-(1-acetyl-piperidin-4-yl)-3-adamantan-1-yl-urea (AR9281) as a potent, selective, and orally available soluble epoxide hydrolase inhibitor with efficacy in rodent models of hypertension and dysglycemia. Bioorg Med Chem Lett21:983-8 (2011) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Bifunctional epoxide hydrolase 2 |
|---|
| Name: | Bifunctional epoxide hydrolase 2 |
|---|
| Synonyms: | Cytosolic epoxide hydrolase 2 | EBifunctional epoxide hydrolase 2 | EPHX2 | Epoxide hydratase | HYES_HUMAN | Lipid-phosphate phosphatase | Soluble epoxide hydrolase (sEH) | epoxide hydrolase 2, cytoplasmic |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 62613.07 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P34913 |
|---|
| Residue: | 555 |
|---|
| Sequence: | MTLRAAVFDLDGVLALPAVFGVLGRTEEALALPRGLLNDAFQKGGPEGATTRLMKGEITL
SQWIPLMEENCRKCSETAKVCLPKNFSIKEIFDKAISARKINRPMLQAALMLRKKGFTTA
ILTNTWLDDRAERDGLAQLMCELKMHFDFLIESCQVGMVKPEPQIYKFLLDTLKASPSEV
VFLDDIGANLKPARDLGMVTILVQDTDTALKELEKVTGIQLLNTPAPLPTSCNPSDMSHG
YVTVKPRVRLHFVELGSGPAVCLCHGFPESWYSWRYQIPALAQAGYRVLAMDMKGYGESS
APPEIEEYCMEVLCKEMVTFLDKLGLSQAVFIGHDWGGMLVWYMALFYPERVRAVASLNT
PFIPANPNMSPLESIKANPVFDYQLYFQEPGVAEAELEQNLSRTFKSLFRASDESVLSMH
KVCEAGGLFVNSPEEPSLSRMVTEEEIQFYVQQFKKSGFRGPLNWYRNMERNWKWACKSL
GRKILIPALMVTAEKDFVLVPQMSQHMEDWIPHLKRGHIEDCGHWTQMDKPTEVNQILIK
WLDSDARNPPVVSKM
|
|
|
|---|
| BDBM50335966 |
|---|
| n/a |
|---|
| Name | BDBM50335966 |
|---|
| Synonyms: | 1-Adamantan-1-yl-3-(1-methanesulfonyl-piperidin-4-yl)-urea | CHEMBL1668935 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C17H29N3O3S |
|---|
| Mol. Mass. | 355.495 |
|---|
| SMILES | CS(=O)(=O)N1CCC(CC1)NC(=O)NC12CC3CC(CC(C3)C1)C2 |TLB:17:18:16.21.15:22,THB:19:18:15:21.20.22,19:20:17.18.23:15,17:16:18.19.23:22| |
|---|
| Structure | 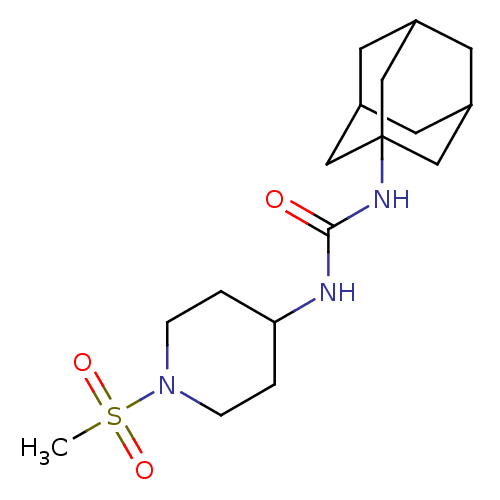
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Anandan, SK; Webb, HK; Chen, D; Wang, YX; Aavula, BR; Cases, S; Cheng, Y; Do, ZN; Mehra, U; Tran, V; Vincelette, J; Waszczuk, J; White, K; Wong, KR; Zhang, LN; Jones, PD; Hammock, BD; Patel, DV; Whitcomb, R; MacIntyre, DE; Sabry, J; Gless, R 1-(1-acetyl-piperidin-4-yl)-3-adamantan-1-yl-urea (AR9281) as a potent, selective, and orally available soluble epoxide hydrolase inhibitor with efficacy in rodent models of hypertension and dysglycemia. Bioorg Med Chem Lett21:983-8 (2011) [PubMed] Article
Anandan, SK; Webb, HK; Chen, D; Wang, YX; Aavula, BR; Cases, S; Cheng, Y; Do, ZN; Mehra, U; Tran, V; Vincelette, J; Waszczuk, J; White, K; Wong, KR; Zhang, LN; Jones, PD; Hammock, BD; Patel, DV; Whitcomb, R; MacIntyre, DE; Sabry, J; Gless, R 1-(1-acetyl-piperidin-4-yl)-3-adamantan-1-yl-urea (AR9281) as a potent, selective, and orally available soluble epoxide hydrolase inhibitor with efficacy in rodent models of hypertension and dysglycemia. Bioorg Med Chem Lett21:983-8 (2011) [PubMed] Article