

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Bile acid receptor | ||
| Ligand | BDBM50336385 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_716785 (CHEMBL1670707) | ||
| EC50 | 2700±n/a nM | ||
| Citation |  Richter, HG; Benson, GM; Bleicher, KH; Blum, D; Chaput, E; Clemann, N; Feng, S; Gardes, C; Grether, U; Hartman, P; Kuhn, B; Martin, RE; Plancher, JM; Rudolph, MG; Schuler, F; Taylor, S Optimization of a novel class of benzimidazole-based farnesoid X receptor (FXR) agonists to improve physicochemical and ADME properties. Bioorg Med Chem Lett21:1134-40 (2011) [PubMed] Article Richter, HG; Benson, GM; Bleicher, KH; Blum, D; Chaput, E; Clemann, N; Feng, S; Gardes, C; Grether, U; Hartman, P; Kuhn, B; Martin, RE; Plancher, JM; Rudolph, MG; Schuler, F; Taylor, S Optimization of a novel class of benzimidazole-based farnesoid X receptor (FXR) agonists to improve physicochemical and ADME properties. Bioorg Med Chem Lett21:1134-40 (2011) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Bile acid receptor | |||
| Name: | Bile acid receptor | ||
| Synonyms: | BAR | Bile acid receptor FXR | FXR | Farnesol receptor HRR-1 | HRR1 | NR1H4 | NR1H4_HUMAN | Nuclear receptor subfamily 1 group H member 4 | RIP14 | RXR-interacting protein 14 | Retinoid X receptor-interacting protein 14 | farnesoid x receptor | ||
| Type: | Nuclear Receptor | ||
| Mol. Mass.: | 55916.24 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q96RI1 | ||
| Residue: | 486 | ||
| Sequence: |
| ||
| BDBM50336385 | |||
| n/a | |||
| Name | BDBM50336385 | ||
| Synonyms: | CHEMBL1668240 | trans-4-((S)-2-(2-(4-chlorophenyl)-5,6-difluoro-1H-benzo[d]imidazol-1-yl)-2-cyclohexylacetamido)cyclohexanecarboxylic acid | ||
| Type | Small organic molecule | ||
| Emp. Form. | C28H30ClF2N3O3 | ||
| Mol. Mass. | 530.006 | ||
| SMILES | OC(=O)[C@H]1CC[C@@H](CC1)NC(=O)[C@H](C1CCCCC1)n1c(nc2cc(F)c(F)cc12)-c1ccc(Cl)cc1 |r,wU:12.20,6.9,wD:3.2,(-9.7,-28.54,;-8.37,-29.32,;-8.39,-30.86,;-7.03,-28.57,;-5.7,-29.35,;-4.35,-28.59,;-4.35,-27.05,;-5.67,-26.27,;-7.01,-27.03,;-3.01,-26.29,;-1.68,-27.07,;-1.69,-28.61,;-.34,-26.31,;.99,-27.09,;.96,-28.63,;2.29,-29.41,;3.63,-28.66,;3.64,-27.12,;2.31,-26.33,;-.33,-24.77,;.57,-23.51,;-.35,-22.26,;-1.82,-22.75,;-3.15,-21.99,;-4.48,-22.76,;-5.82,-21.99,;-4.48,-24.3,;-5.82,-25.07,;-3.15,-25.07,;-1.81,-24.3,;2.11,-23.5,;2.89,-24.83,;4.42,-24.82,;5.19,-23.48,;6.73,-23.47,;4.4,-22.15,;2.87,-22.16,)| | ||
| Structure | 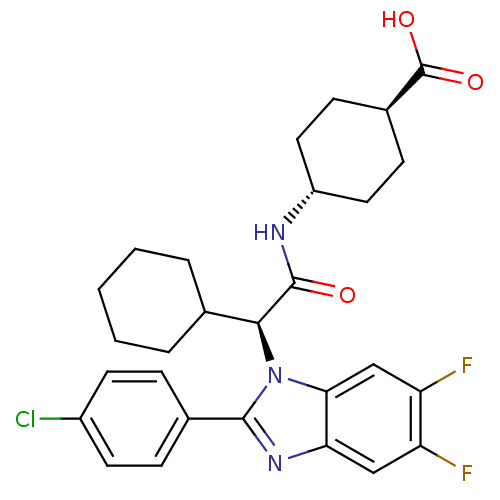
| ||