

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Cytochrome P450 3A4 | ||
| Ligand | BDBM50336317 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_730102 (CHEMBL1695868) | ||
| IC50 | >25000±n/a nM | ||
| Citation |  Jin, M; Gokhale, PC; Cooke, A; Foreman, K; Buck, E; May, EW; Feng, L; Bittner, MA; Kadalbajoo, M; Landfair, D; Siu, KW; Stolz, KM; Werner, DS; Laufer, RS; Li, AH; Dong, H; Steinig, AG; Kleinberg, A; Yao, Y; Pachter, JA; Wild, R; Mulvihill, MJ Discovery of an Orally Efficacious Imidazo[5,1-f][1,2,4]triazine Dual Inhibitor of IGF-1R and IR. ACS Med Chem Lett1:510-515 (2010) [PubMed] Article Jin, M; Gokhale, PC; Cooke, A; Foreman, K; Buck, E; May, EW; Feng, L; Bittner, MA; Kadalbajoo, M; Landfair, D; Siu, KW; Stolz, KM; Werner, DS; Laufer, RS; Li, AH; Dong, H; Steinig, AG; Kleinberg, A; Yao, Y; Pachter, JA; Wild, R; Mulvihill, MJ Discovery of an Orally Efficacious Imidazo[5,1-f][1,2,4]triazine Dual Inhibitor of IGF-1R and IR. ACS Med Chem Lett1:510-515 (2010) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Cytochrome P450 3A4 | |||
| Name: | Cytochrome P450 3A4 | ||
| Synonyms: | Albendazole monooxygenase | Albendazole sulfoxidase | CP3A4_HUMAN | CYP3A3 | CYP3A4 | CYPIIIA3 | CYPIIIA4 | Cytochrome P450 3A3 | Cytochrome P450 3A4 (CYP3A4) | Cytochrome P450 HLp | Nifedipine oxidase | Quinine 3-monooxygenase | Taurochenodeoxycholate 6-alpha-hydroxylase | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 57349.57 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | n/a | ||
| Residue: | 503 | ||
| Sequence: |
| ||
| BDBM50336317 | |||
| n/a | |||
| Name | BDBM50336317 | ||
| Synonyms: | CHEMBL1667935 | cis-3-[4-Amino-5-(8-fluoro-2-phenylquinolin-7-yl)imidazo[5,1-f][1,2,4]triazin-7-yl]-1-methylcyclobutanol | cis-3-[4-amino-5-(8-fluoro-2-phenylquinolin-7-yl)-imidazo[5,1-f][1,2,4]triazin-7-yl]-1-methyl-cyclobutanol | ||
| Type | Small organic molecule | ||
| Emp. Form. | C25H21FN6O | ||
| Mol. Mass. | 440.4722 | ||
| SMILES | C[C@@]1(O)C[C@@H](C1)c1nc(-c2ccc3ccc(nc3c2F)-c2ccccc2)c2c(N)ncnn12 |r,wU:4.6,1.1,wD:1.0,(-2.6,-22.44,;-3.93,-21.67,;-4.34,-23.15,;-3.24,-20.29,;-4.61,-19.61,;-5.31,-20.98,;-5.1,-18.14,;-4.2,-16.89,;-5.11,-15.65,;-4.34,-14.32,;-5.11,-12.99,;-4.34,-11.66,;-2.79,-11.65,;-2.03,-10.33,;-.5,-10.33,;.27,-11.66,;-.5,-12.99,;-2.03,-12.99,;-2.8,-14.32,;-2.03,-15.66,;1.81,-11.66,;2.58,-13,;4.12,-13,;4.89,-11.66,;4.11,-10.32,;2.57,-10.33,;-6.58,-16.13,;-7.91,-15.37,;-7.92,-13.83,;-9.24,-16.14,;-9.25,-17.68,;-7.91,-18.46,;-6.57,-17.68,)| | ||
| Structure | 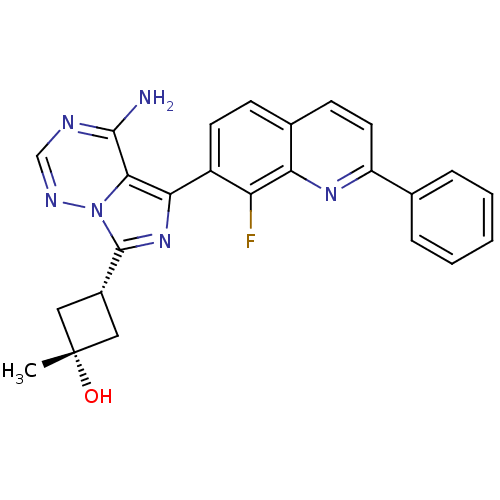
| ||