| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Glycogen phosphorylase, muscle form |
|---|
| Ligand | BDBM50346688 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_753234 (CHEMBL1799389) |
|---|
| IC50 | 2230±n/a nM |
|---|
| Citation |  Liang, Z; Zhang, L; Li, L; Liu, J; Li, H; Zhang, L; Chen, L; Cheng, K; Zheng, M; Wen, X; Zhang, P; Hao, J; Gong, Y; Zhang, X; Zhu, X; Chen, J; Liu, H; Jiang, H; Luo, C; Sun, H Identification of pentacyclic triterpenes derivatives as potent inhibitors against glycogen phosphorylase based on 3D-QSAR studies. Eur J Med Chem46:2011-21 (2011) [PubMed] Article Liang, Z; Zhang, L; Li, L; Liu, J; Li, H; Zhang, L; Chen, L; Cheng, K; Zheng, M; Wen, X; Zhang, P; Hao, J; Gong, Y; Zhang, X; Zhu, X; Chen, J; Liu, H; Jiang, H; Luo, C; Sun, H Identification of pentacyclic triterpenes derivatives as potent inhibitors against glycogen phosphorylase based on 3D-QSAR studies. Eur J Med Chem46:2011-21 (2011) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Glycogen phosphorylase, muscle form |
|---|
| Name: | Glycogen phosphorylase, muscle form |
|---|
| Synonyms: | Glycogen Phosphorylase (PYGM) | Glycogen phosphorylase a (RMGPa) | Glycogen phosphorylase, muscle form | Myophosphorylase | PYGM | PYGM_RABIT |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 97296.32 |
|---|
| Organism: | Oryctolagus cuniculus (rabbit) |
|---|
| Description: | Phosphorylation of Ser-15 converts phosphorylase B (unphosphorylated) to phosphorylase A. |
|---|
| Residue: | 843 |
|---|
| Sequence: | MSRPLSDQEKRKQISVRGLAGVENVTELKKNFNRHLHFTLVKDRNVATPRDYYFALAHTV
RDHLVGRWIRTQQHYYEKDPKRIYYLSLEFYMGRTLQNTMVNLALENACDEATYQLGLDM
EELEEIEEDAGLGNGGLGRLAACFLDSMATLGLAAYGYGIRYEFGIFNQKICGGWQMEEA
DDWLRYGNPWEKARPEFTLPVHFYGRVEHTSQGAKWVDTQVVLAMPYDTPVPGYRNNVVN
TMRLWSAKAPNDFNLKDFNVGGYIQAVLDRNLAENISRVLYPNDNFFEGKELRLKQEYFV
VAATLQDIIRRFKSSKFGCRDPVRTNFDAFPDKVAIQLNDTHPSLAIPELMRVLVDLERL
DWDKAWEVTVKTCAYTNHTVLPEALERWPVHLLETLLPRHLQIIYEINQRFLNRVAAAFP
GDVDRLRRMSLVEEGAVKRINMAHLCIAGSHAVNGVARIHSEILKKTIFKDFYELEPHKF
QNKTNGITPRRWLVLCNPGLAEIIAERIGEEYISDLDQLRKLLSYVDDEAFIRDVAKVKQ
ENKLKFAAYLEREYKVHINPNSLFDVQVKRIHEYKRQLLNCLHVITLYNRIKKEPNKFVV
PRTVMIGGKAAPGYHMAKMIIKLITAIGDVVNHDPVVGDRLRVIFLENYRVSLAEKVIPA
ADLSEQISTAGTEASGTGNMKFMLNGALTIGTMDGANVEMAEEAGEENFFIFGMRVEDVD
RLDQRGYNAQEYYDRIPELRQIIEQLSSGFFSPKQPDLFKDIVNMLMHHDRFKVFADYEE
YVKCQERVSALYKNPREWTRMVIRNIATSGKFSSDRTIAQYAREIWGVEPSRQRLPAPDE
KIP
|
|
|
|---|
| BDBM50346688 |
|---|
| n/a |
|---|
| Name | BDBM50346688 |
|---|
| Synonyms: | CHEMBL1796878 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C38H56O9 |
|---|
| Mol. Mass. | 656.8458 |
|---|
| SMILES | CC(=O)O[C@H]1CC[C@@]2(C)[C@@H](CC[C@]3(C)[C@@H]2CC=C2[C@@H]4CC(C)(C)CC[C@@]4(CC[C@@]32C)C(=O)OC[C@H](O)[C@H]2OC(=O)C(O)C2=O)C1(C)C |r,t:17| |
|---|
| Structure | 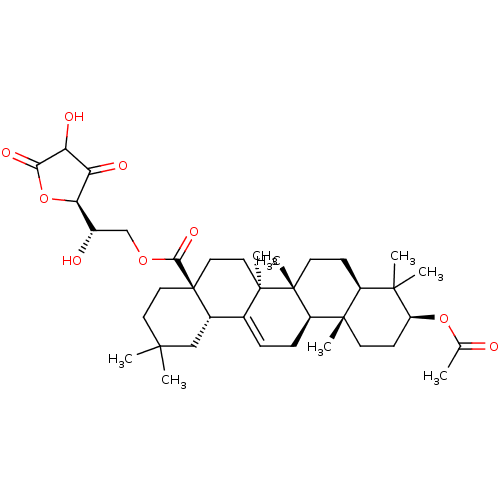
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Liang, Z; Zhang, L; Li, L; Liu, J; Li, H; Zhang, L; Chen, L; Cheng, K; Zheng, M; Wen, X; Zhang, P; Hao, J; Gong, Y; Zhang, X; Zhu, X; Chen, J; Liu, H; Jiang, H; Luo, C; Sun, H Identification of pentacyclic triterpenes derivatives as potent inhibitors against glycogen phosphorylase based on 3D-QSAR studies. Eur J Med Chem46:2011-21 (2011) [PubMed] Article
Liang, Z; Zhang, L; Li, L; Liu, J; Li, H; Zhang, L; Chen, L; Cheng, K; Zheng, M; Wen, X; Zhang, P; Hao, J; Gong, Y; Zhang, X; Zhu, X; Chen, J; Liu, H; Jiang, H; Luo, C; Sun, H Identification of pentacyclic triterpenes derivatives as potent inhibitors against glycogen phosphorylase based on 3D-QSAR studies. Eur J Med Chem46:2011-21 (2011) [PubMed] Article