| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Dihydroorotate dehydrogenase (quinone), mitochondrial |
|---|
| Ligand | BDBM50365240 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_808186 (CHEMBL1961098) |
|---|
| IC50 | >100000±n/a nM |
|---|
| Citation |  Coteron, JM; Marco, M; Esquivias, J; Deng, X; White, KL; White, J; Koltun, M; El Mazouni, F; Kokkonda, S; Katneni, K; Bhamidipati, R; Shackleford, DM; Angulo-Barturen, I; Ferrer, SB; Jiménez-Díaz, MB; Gamo, FJ; Goldsmith, EJ; Charman, WN; Bathurst, I; Floyd, D; Matthews, D; Burrows, JN; Rathod, PK; Charman, SA; Phillips, MA Structure-guided lead optimization of triazolopyrimidine-ring substituents identifies potent Plasmodium falciparum dihydroorotate dehydrogenase inhibitors with clinical candidate potential. J Med Chem54:5540-61 (2011) [PubMed] Article Coteron, JM; Marco, M; Esquivias, J; Deng, X; White, KL; White, J; Koltun, M; El Mazouni, F; Kokkonda, S; Katneni, K; Bhamidipati, R; Shackleford, DM; Angulo-Barturen, I; Ferrer, SB; Jiménez-Díaz, MB; Gamo, FJ; Goldsmith, EJ; Charman, WN; Bathurst, I; Floyd, D; Matthews, D; Burrows, JN; Rathod, PK; Charman, SA; Phillips, MA Structure-guided lead optimization of triazolopyrimidine-ring substituents identifies potent Plasmodium falciparum dihydroorotate dehydrogenase inhibitors with clinical candidate potential. J Med Chem54:5540-61 (2011) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Dihydroorotate dehydrogenase (quinone), mitochondrial |
|---|
| Name: | Dihydroorotate dehydrogenase (quinone), mitochondrial |
|---|
| Synonyms: | DHODH | DHOdehase | Dihydroorotate dehydrogenase | Dihydroorotate dehydrogenase (DHODH) | Dihydroorotate dehydrogenase, mitochondrial | Dihydroorotate oxidase | Dihydroorotate oxidase (DHODH) | PYRD_HUMAN |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 42881.33 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q02127 |
|---|
| Residue: | 395 |
|---|
| Sequence: | MAWRHLKKRAQDAVIILGGGGLLFASYLMATGDERFYAEHLMPTLQGLLDPESAHRLAVR
FTSLGLLPRARFQDSDMLEVRVLGHKFRNPVGIAAGFDKHGEAVDGLYKMGFGFVEIGSV
TPKPQEGNPRPRVFRLPEDQAVINRYGFNSHGLSVVEHRLRARQQKQAKLTEDGLPLGVN
LGKNKTSVDAAEDYAEGVRVLGPLADYLVVNVSSPNTAGLRSLQGKAELRRLLTKVLQER
DGLRRVHRPAVLVKIAPDLTSQDKEDIASVVKELGIDGLIVTNTTVSRPAGLQGALRSET
GGLSGKPLRDLSTQTIREMYALTQGRVPIIGVGGVSSGQDALEKIRAGASLVQLYTALTF
WGPPVVGKVKRELEALLKEQGFGGVTDAIGADHRR
|
|
|
|---|
| BDBM50365240 |
|---|
| n/a |
|---|
| Name | BDBM50365240 |
|---|
| Synonyms: | CHEMBL1956272 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C17H18F3N5 |
|---|
| Mol. Mass. | 349.3535 |
|---|
| SMILES | Cc1cc(Nc2ccc(cc2)C(F)(F)F)n2nc(nc2n1)C(C)(C)C |
|---|
| Structure | 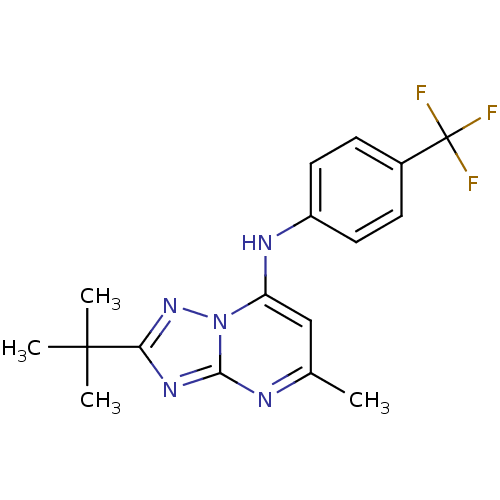
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Coteron, JM; Marco, M; Esquivias, J; Deng, X; White, KL; White, J; Koltun, M; El Mazouni, F; Kokkonda, S; Katneni, K; Bhamidipati, R; Shackleford, DM; Angulo-Barturen, I; Ferrer, SB; Jiménez-Díaz, MB; Gamo, FJ; Goldsmith, EJ; Charman, WN; Bathurst, I; Floyd, D; Matthews, D; Burrows, JN; Rathod, PK; Charman, SA; Phillips, MA Structure-guided lead optimization of triazolopyrimidine-ring substituents identifies potent Plasmodium falciparum dihydroorotate dehydrogenase inhibitors with clinical candidate potential. J Med Chem54:5540-61 (2011) [PubMed] Article
Coteron, JM; Marco, M; Esquivias, J; Deng, X; White, KL; White, J; Koltun, M; El Mazouni, F; Kokkonda, S; Katneni, K; Bhamidipati, R; Shackleford, DM; Angulo-Barturen, I; Ferrer, SB; Jiménez-Díaz, MB; Gamo, FJ; Goldsmith, EJ; Charman, WN; Bathurst, I; Floyd, D; Matthews, D; Burrows, JN; Rathod, PK; Charman, SA; Phillips, MA Structure-guided lead optimization of triazolopyrimidine-ring substituents identifies potent Plasmodium falciparum dihydroorotate dehydrogenase inhibitors with clinical candidate potential. J Med Chem54:5540-61 (2011) [PubMed] Article