| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform |
|---|
| Ligand | BDBM50396628 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_864130 (CHEMBL2174978) |
|---|
| IC50 | 547±n/a nM |
|---|
| Citation |  Murray, JM; Sweeney, ZK; Chan, BK; Balazs, M; Bradley, E; Castanedo, G; Chabot, C; Chantry, D; Flagella, M; Goldstein, DM; Kondru, R; Lesnick, J; Li, J; Lucas, MC; Nonomiya, J; Pang, J; Price, S; Salphati, L; Safina, B; Savy, PP; Seward, EM; Ultsch, M; Sutherlin, DP Potent and highly selective benzimidazole inhibitors of PI3-kinase delta. J Med Chem55:7686-95 (2012) [PubMed] Article Murray, JM; Sweeney, ZK; Chan, BK; Balazs, M; Bradley, E; Castanedo, G; Chabot, C; Chantry, D; Flagella, M; Goldstein, DM; Kondru, R; Lesnick, J; Li, J; Lucas, MC; Nonomiya, J; Pang, J; Price, S; Salphati, L; Safina, B; Savy, PP; Seward, EM; Ultsch, M; Sutherlin, DP Potent and highly selective benzimidazole inhibitors of PI3-kinase delta. J Med Chem55:7686-95 (2012) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform |
|---|
| Name: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform |
|---|
| Synonyms: | PI3-kinase p110 subunit beta | PI3-kinase subunit p110-beta | PI3Kbeta | PIK3C1 | PIK3CB | PK3CB_HUMAN | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta (PI3Kbeta) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform (PI3K beta) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform (PI3K) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform (PI3K-beta) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform (PI3Kbeta) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform (PI3Kÿ²) | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform | Phosphoinositide 3-Kinase (PI3K), beta | Phosphoinositide 3-Kinase (PI3K), beta Chain A | Phosphoinositide-3-kinase (PI3K beta) | PtdIns-3-kinase p110 |
|---|
| Type: | Enzyme Subunit |
|---|
| Mol. Mass.: | 122769.00 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P42338 |
|---|
| Residue: | 1070 |
|---|
| Sequence: | MCFSFIMPPAMADILDIWAVDSQIASDGSIPVDFLLPTGIYIQLEVPREATISYIKQMLW
KQVHNYPMFNLLMDIDSYMFACVNQTAVYEELEDETRRLCDVRPFLPVLKLVTRSCDPGE
KLDSKIGVLIGKGLHEFDSLKDPEVNEFRRKMRKFSEEKILSLVGLSWMDWLKQTYPPEH
EPSIPENLEDKLYGGKLIVAVHFENCQDVFSFQVSPNMNPIKVNELAIQKRLTIHGKEDE
VSPYDYVLQVSGRVEYVFGDHPLIQFQYIRNCVMNRALPHFILVECCKIKKMYEQEMIAI
EAAINRNSSNLPLPLPPKKTRIISHVWENNNPFQIVLVKGNKLNTEETVKVHVRAGLFHG
TELLCKTIVSSEVSGKNDHIWNEPLEFDINICDLPRMARLCFAVYAVLDKVKTKKSTKTI
NPSKYQTIRKAGKVHYPVAWVNTMVFDFKGQLRTGDIILHSWSSFPDELEEMLNPMGTVQ
TNPYTENATALHVKFPENKKQPYYYPPFDKIIEKAAEIASSDSANVSSRGGKKFLPVLKE
ILDRDPLSQLCENEMDLIWTLRQDCREIFPQSLPKLLLSIKWNKLEDVAQLQALLQIWPK
LPPREALELLDFNYPDQYVREYAVGCLRQMSDEELSQYLLQLVQVLKYEPFLDCALSRFL
LERALGNRRIGQFLFWHLRSEVHIPAVSVQFGVILEAYCRGSVGHMKVLSKQVEALNKLK
TLNSLIKLNAVKLNRAKGKEAMHTCLKQSAYREALSDLQSPLNPCVILSELYVEKCKYMD
SKMKPLWLVYNNKVFGEDSVGVIFKNGDDLRQDMLTLQMLRLMDLLWKEAGLDLRMLPYG
CLATGDRSGLIEVVSTSETIADIQLNSSNVAAAAAFNKDALLNWLKEYNSGDDLDRAIEE
FTLSCAGYCVASYVLGIGDRHSDNIMVKKTGQLFHIDFGHILGNFKSKFGIKRERVPFIL
TYDFIHVIQQGKTGNTEKFGRFRQCCEDAYLILRRHGNLFITLFALMLTAGLPELTSVKD
IQYLKDSLALGKSEEEALKQFKQKFDEALRESWTTKVNWMAHTVRKDYRS
|
|
|
|---|
| BDBM50396628 |
|---|
| n/a |
|---|
| Name | BDBM50396628 |
|---|
| Synonyms: | CHEMBL2171944 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C28H38N8O2 |
|---|
| Mol. Mass. | 518.6537 |
|---|
| SMILES | CCc1nc2ccccc2n1-c1nc(N2CCOCC2)c2nc(CN3CCC(CC3)C(C)(C)O)n(C)c2n1 |
|---|
| Structure | 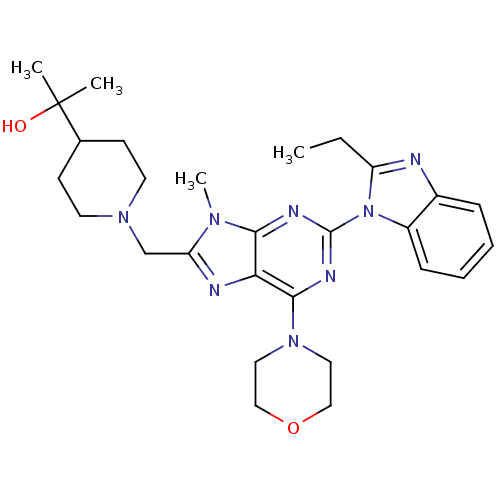
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Murray, JM; Sweeney, ZK; Chan, BK; Balazs, M; Bradley, E; Castanedo, G; Chabot, C; Chantry, D; Flagella, M; Goldstein, DM; Kondru, R; Lesnick, J; Li, J; Lucas, MC; Nonomiya, J; Pang, J; Price, S; Salphati, L; Safina, B; Savy, PP; Seward, EM; Ultsch, M; Sutherlin, DP Potent and highly selective benzimidazole inhibitors of PI3-kinase delta. J Med Chem55:7686-95 (2012) [PubMed] Article
Murray, JM; Sweeney, ZK; Chan, BK; Balazs, M; Bradley, E; Castanedo, G; Chabot, C; Chantry, D; Flagella, M; Goldstein, DM; Kondru, R; Lesnick, J; Li, J; Lucas, MC; Nonomiya, J; Pang, J; Price, S; Salphati, L; Safina, B; Savy, PP; Seward, EM; Ultsch, M; Sutherlin, DP Potent and highly selective benzimidazole inhibitors of PI3-kinase delta. J Med Chem55:7686-95 (2012) [PubMed] Article