

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Peroxisome proliferator-activated receptor gamma | ||
| Ligand | BDBM50002871 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_438960 (CHEMBL889304) | ||
| Ki | 309±n/a nM | ||
| Citation |  Trump, RP; Cobb, JE; Shearer, BG; Lambert, MH; Nolte, RT; Willson, TM; Buckholz, RG; Zhao, SM; Leesnitzer, LM; Iannone, MA; Pearce, KH; Billin, AN; Hoekstra, WJ Co-crystal structure guided array synthesis of PPARgamma inverse agonists. Bioorg Med Chem Lett17:3916-20 (2007) [PubMed] Article Trump, RP; Cobb, JE; Shearer, BG; Lambert, MH; Nolte, RT; Willson, TM; Buckholz, RG; Zhao, SM; Leesnitzer, LM; Iannone, MA; Pearce, KH; Billin, AN; Hoekstra, WJ Co-crystal structure guided array synthesis of PPARgamma inverse agonists. Bioorg Med Chem Lett17:3916-20 (2007) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Peroxisome proliferator-activated receptor gamma | |||
| Name: | Peroxisome proliferator-activated receptor gamma | ||
| Synonyms: | NR1C3 | Nuclear receptor subfamily 1 group C member 3 | PPAR-gamma | PPARG | PPARG_HUMAN | Peroxisome proliferator-activated receptor | Peroxisome proliferator-activated receptor gamma (PPAR gamma) | Peroxisome proliferator-activated receptor gamma (PPARG) | Peroxisome proliferator-activated receptor gamma (PPARγ) | Peroxisome proliferator-activated receptor gamma/Nuclear receptor corepressor 2 | peroxisome proliferator-activated receptor gamma isoform 2 | ||
| Type: | Nuclear Receptor | ||
| Mol. Mass.: | 57613.46 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P37231 | ||
| Residue: | 505 | ||
| Sequence: |
| ||
| BDBM50002871 | |||
| n/a | |||
| Name | BDBM50002871 | ||
| Synonyms: | CHEMBL396987 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C41H43N3O5 | ||
| Mol. Mass. | 657.7972 | ||
| SMILES | Cc1oc(nc1CCOc1ccc(C[C@@H](CNC(=O)C2CCC(O)CC2)Nc2ccccc2C(=O)c2ccccc2)cc1)-c1ccccc1 |wU:14.27,(2.33,-4.31,;1.09,-5.24,;-.38,-4.78,;-1.27,-6.05,;-.34,-7.28,;1.12,-6.78,;2.44,-7.56,;3.79,-6.81,;5.11,-7.6,;6.45,-6.84,;7.77,-7.63,;9.12,-6.88,;9.14,-5.34,;10.48,-4.58,;11.8,-5.37,;13.15,-4.61,;14.47,-5.4,;15.81,-4.64,;15.83,-3.1,;17.14,-5.43,;17.11,-6.96,;18.43,-7.75,;19.78,-7,;21.1,-7.79,;19.8,-5.46,;18.47,-4.66,;11.79,-6.91,;13.11,-7.69,;14.45,-6.93,;15.77,-7.71,;15.76,-9.26,;14.42,-10.01,;13.09,-9.23,;11.75,-9.98,;10.42,-9.2,;11.73,-11.52,;10.39,-12.27,;10.37,-13.81,;11.69,-14.6,;13.04,-13.84,;13.05,-12.3,;7.8,-4.55,;6.47,-5.31,;-2.81,-6.07,;-3.6,-4.75,;-5.14,-4.77,;-5.89,-6.12,;-5.11,-7.44,;-3.56,-7.42,)| | ||
| Structure | 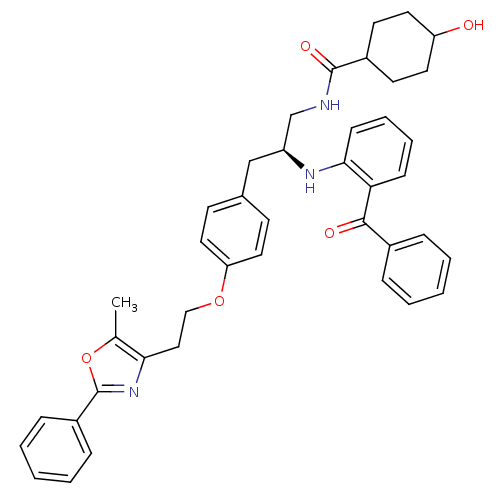
| ||