

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Mu-type opioid receptor | ||
| Ligand | BDBM50101091 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1452451 (CHEMBL3362475) | ||
| Ki | 9.0±n/a nM | ||
| Citation |  Schunk, S; Linz, K; Frormann, S; Hinze, C; Oberbörsch, S; Sundermann, B; Zemolka, S; Englberger, W; Germann, T; Christoph, T; Kögel, BY; Schröder, W; Harlfinger, S; Saunders, D; Kless, A; Schick, H; Sonnenschein, H Discovery of Spiro[cyclohexane-dihydropyrano[3,4-b]indole]-amines as Potent NOP and Opioid Receptor Agonists. ACS Med Chem Lett5:851-6 (2014) [PubMed] Article Schunk, S; Linz, K; Frormann, S; Hinze, C; Oberbörsch, S; Sundermann, B; Zemolka, S; Englberger, W; Germann, T; Christoph, T; Kögel, BY; Schröder, W; Harlfinger, S; Saunders, D; Kless, A; Schick, H; Sonnenschein, H Discovery of Spiro[cyclohexane-dihydropyrano[3,4-b]indole]-amines as Potent NOP and Opioid Receptor Agonists. ACS Med Chem Lett5:851-6 (2014) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Mu-type opioid receptor | |||
| Name: | Mu-type opioid receptor | ||
| Synonyms: | M-OR-1 | MOP | MOR-1 | MOR1 | MUOR1 | Mu Opioid Receptor | Mu opiate receptor | OPIATE Mu | OPRM1 | OPRM_HUMAN | hMOP | mu-type opioid receptor isoform MOR-1 | ||
| Type: | G Protein-Coupled Receptor (GPCR) | ||
| Mol. Mass.: | 44789.51 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P35372 | ||
| Residue: | 400 | ||
| Sequence: |
| ||
| BDBM50101091 | |||
| n/a | |||
| Name | BDBM50101091 | ||
| Synonyms: | CHEMBL3326223 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C24H32Cl2FN3 | ||
| Mol. Mass. | 452.435 | ||
| SMILES | Cl.Cl.CN(C)[C@]1(CC[C@@H](CC1)NCCc1c[nH]c2ccc(F)cc12)c1ccccc1 |r,wU:5.2,wD:8.9,(50.15,-23.7,;44.1,-26.45,;47.35,-21.61,;46.01,-20.85,;46.01,-19.3,;44.68,-21.62,;43.91,-22.95,;42.37,-22.95,;41.6,-21.63,;42.36,-20.29,;43.9,-20.29,;40.06,-21.63,;39.26,-22.95,;37.72,-22.91,;36.92,-24.23,;37.52,-25.66,;36.35,-26.66,;35.03,-25.86,;33.57,-26.31,;32.44,-25.27,;32.79,-23.76,;31.66,-22.71,;34.26,-23.31,;35.38,-24.36,;45.47,-22.94,;44.73,-24.28,;45.53,-25.6,;47.07,-25.57,;47.81,-24.21,;47.01,-22.9,)| | ||
| Structure | 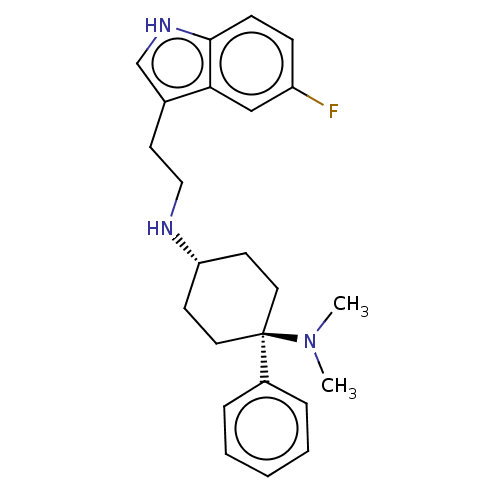
| ||