| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Lysine-specific demethylase 5C |
|---|
| Ligand | BDBM50153099 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1561972 (CHEMBL3778979) |
|---|
| IC50 | 24±n/a nM |
|---|
| Citation |  Bavetsias, V; Lanigan, RM; Ruda, GF; Atrash, B; McLaughlin, MG; Tumber, A; Mok, NY; Le Bihan, YV; Dempster, S; Boxall, KJ; Jeganathan, F; Hatch, SB; Savitsky, P; Velupillai, S; Krojer, T; England, KS; Sejberg, J; Thai, C; Donovan, A; Pal, A; Scozzafava, G; Bennett, JM; Kawamura, A; Johansson, C; Szykowska, A; Gileadi, C; Burgess-Brown, NA; von Delft, F; Oppermann, U; Walters, Z; Shipley, J; Raynaud, FI; Westaway, SM; Prinjha, RK; Fedorov, O; Burke, R; Schofield, CJ; Westwood, IM; Bountra, C; Müller, S; van Montfort, RL; Brennan, PE; Blagg, J 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors. J Med Chem59:1388-409 (2016) [PubMed] Article Bavetsias, V; Lanigan, RM; Ruda, GF; Atrash, B; McLaughlin, MG; Tumber, A; Mok, NY; Le Bihan, YV; Dempster, S; Boxall, KJ; Jeganathan, F; Hatch, SB; Savitsky, P; Velupillai, S; Krojer, T; England, KS; Sejberg, J; Thai, C; Donovan, A; Pal, A; Scozzafava, G; Bennett, JM; Kawamura, A; Johansson, C; Szykowska, A; Gileadi, C; Burgess-Brown, NA; von Delft, F; Oppermann, U; Walters, Z; Shipley, J; Raynaud, FI; Westaway, SM; Prinjha, RK; Fedorov, O; Burke, R; Schofield, CJ; Westwood, IM; Bountra, C; Müller, S; van Montfort, RL; Brennan, PE; Blagg, J 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors. J Med Chem59:1388-409 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Lysine-specific demethylase 5C |
|---|
| Name: | Lysine-specific demethylase 5C |
|---|
| Synonyms: | DXS1272E | Histone demethylase JARID1C | JARID1C | Jumonji/ARID domain-containing protein 1C | KDM5C | KDM5C_HUMAN | Lysine-specific demethylase 5C (KDM5C) | Lysine-specific demethylase 5C (KDM5Flag-5C-FL) | Protein SmcX | Protein Xe169 | SMCX | XE169 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 175691.17 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P41229 |
|---|
| Residue: | 1560 |
|---|
| Sequence: | MEPGSDDFLPPPECPVFEPSWAEFRDPLGYIAKIRPIAEKSGICKIRPPADWQPPFAVEV
DNFRFTPRIQRLNELEAQTRVKLNYLDQIAKFWEIQGSSLKIPNVERRILDLYSLSKIVV
EEGGYEAICKDRRWARVAQRLNYPPGKNIGSLLRSHYERIVYPYEMYQSGANLVQCNTRP
FDNEEKDKEYKPHSIPLRQSVQPSKFNSYGRRAKRLQPDPEPTEEDIEKNPELKKLQIYG
AGPKMMGLGLMAKDKTLRKKDKEGPECPPTVVVKEELGGDVKVESTSPKTFLESKEELSH
SPEPCTKMTMRLRRNHSNAQFIESYVCRMCSRGDEDDKLLLCDGCDDNYHIFCLLPPLPE
IPKGVWRCPKCVMAECKRPPEAFGFEQATREYTLQSFGEMADSFKADYFNMPVHMVPTEL
VEKEFWRLVNSIEEDVTVEYGADIHSKEFGSGFPVSDSKRHLTPEEEEYATSGWNLNVMP
VLEQSVLCHINADISGMKVPWLYVGMVFSAFCWHIEDHWSYSINYLHWGEPKTWYGVPSL
AAEHLEEVMKKLTPELFDSQPDLLHQLVTLMNPNTLMSHGVPVVRTNQCAGEFVITFPRA
YHSGFNQGYNFAEAVNFCTADWLPAGRQCIEHYRRLRRYCVFSHEELICKMAACPEKLDL
NLAAAVHKEMFIMVQEERRLRKALLEKGITEAEREAFELLPDDERQCIKCKTTCFLSALA
CYDCPDGLVCLSHINDLCKCSSSRQYLRYRYTLDELPAMLHKLKVRAESFDTWANKVRVA
LEVEDGRKRSLEELRALESEARERRFPNSELLQQLKNCLSEAEACVSRALGLVSGQEAGP
HRVAGLQMTLTELRAFLDQMNNLPCAMHQIGDVKGVLEQVEAYQAEAREALASLPSSPGL
LQSLLERGRQLGVEVPEAQQLQRQVEQARWLDEVKRTLAPSARRGTLAVMRGLLVAGASV
APSPAVDKAQAELQELLTIAERWEEKAHLCLEARQKHPPATLEAIIREAENIPVHLPNIQ
ALKEALAKARAWIADVDEIQNGDHYPCLDDLEGLVAVGRDLPVGLEELRQLELQVLTAHS
WREKASKTFLKKNSCYTLLEVLCPCADAGSDSTKRSRWMEKELGLYKSDTELLGLSAQDL
RDPGSVIVAFKEGEQKEKEGILQLRRTNSAKPSPLASSSTASSTTSICVCGQVLAGAGAL
QCDLCQDWFHGRCVSVPRLLSSPRPNPTSSPLLAWWEWDTKFLCPLCMRSRRPRLETILA
LLVALQRLPVRLPEGEALQCLTERAISWQGRARQALASEDVTALLGRLAELRQRLQAEPR
PEEPPNYPAAPASDPLREGSGKDMPKVQGLLENGDSVTSPEKVAPEEGSGKRDLELLSSL
LPQLTGPVLELPEATRAPLEELMMEGDLLEVTLDENHSIWQLLQAGQPPDLERIRTLLEL
EKAERHGSRARGRALERRRRRKVDRGGEGDDPAREELEPKRVRSSGPEAEEVQEEEELEE
ETGGEGPPAPIPTTGSPSTQENQNGLEPAEGTTSGPSAPFSTLTPRLHLPCPQQPPQQQL
|
|
|
|---|
| BDBM50153099 |
|---|
| n/a |
|---|
| Name | BDBM50153099 |
|---|
| Synonyms: | CHEMBL3775814 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C24H23N7O |
|---|
| Mol. Mass. | 425.4857 |
|---|
| SMILES | O=c1[nH]cnc2c(nccc12)-n1cc(CCN2CCC(CC2)c2ccc(cc2)C#N)cn1 |
|---|
| Structure | 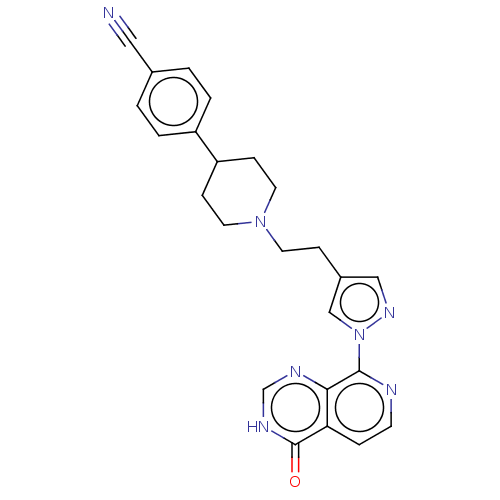
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Bavetsias, V; Lanigan, RM; Ruda, GF; Atrash, B; McLaughlin, MG; Tumber, A; Mok, NY; Le Bihan, YV; Dempster, S; Boxall, KJ; Jeganathan, F; Hatch, SB; Savitsky, P; Velupillai, S; Krojer, T; England, KS; Sejberg, J; Thai, C; Donovan, A; Pal, A; Scozzafava, G; Bennett, JM; Kawamura, A; Johansson, C; Szykowska, A; Gileadi, C; Burgess-Brown, NA; von Delft, F; Oppermann, U; Walters, Z; Shipley, J; Raynaud, FI; Westaway, SM; Prinjha, RK; Fedorov, O; Burke, R; Schofield, CJ; Westwood, IM; Bountra, C; Müller, S; van Montfort, RL; Brennan, PE; Blagg, J 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors. J Med Chem59:1388-409 (2016) [PubMed] Article
Bavetsias, V; Lanigan, RM; Ruda, GF; Atrash, B; McLaughlin, MG; Tumber, A; Mok, NY; Le Bihan, YV; Dempster, S; Boxall, KJ; Jeganathan, F; Hatch, SB; Savitsky, P; Velupillai, S; Krojer, T; England, KS; Sejberg, J; Thai, C; Donovan, A; Pal, A; Scozzafava, G; Bennett, JM; Kawamura, A; Johansson, C; Szykowska, A; Gileadi, C; Burgess-Brown, NA; von Delft, F; Oppermann, U; Walters, Z; Shipley, J; Raynaud, FI; Westaway, SM; Prinjha, RK; Fedorov, O; Burke, R; Schofield, CJ; Westwood, IM; Bountra, C; Müller, S; van Montfort, RL; Brennan, PE; Blagg, J 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors. J Med Chem59:1388-409 (2016) [PubMed] Article