| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Heat shock protein HSP 90-alpha |
|---|
| Ligand | BDBM50210933 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1635598 (CHEMBL3878496) |
|---|
| IC50 | 50±n/a nM |
|---|
| Citation |  Jeong, JH; Oh, YJ; Lho, Y; Park, SY; Liu, KH; Ha, E; Seo, YH Targeting the entry region of Hsp90's ATP binding pocket with a novel 6,7-dihydrothieno[3,2-c]pyridin-5(4H)-yl amide. Eur J Med Chem124:1069-1080 (2016) [PubMed] Article Jeong, JH; Oh, YJ; Lho, Y; Park, SY; Liu, KH; Ha, E; Seo, YH Targeting the entry region of Hsp90's ATP binding pocket with a novel 6,7-dihydrothieno[3,2-c]pyridin-5(4H)-yl amide. Eur J Med Chem124:1069-1080 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Heat shock protein HSP 90-alpha |
|---|
| Name: | Heat shock protein HSP 90-alpha |
|---|
| Synonyms: | HS90A_HUMAN | HSP 86 | HSP86 | HSP90A | HSP90AA1 | HSPC1 | HSPCA | Heat Shock Protein 90 (Hsp90) | Heat shock 86 kDa | Heat shock protein HSP 90 (HSP90) | Heat shock protein HSP 90-alpha (HSP90) | Heat shock protein HSP 90-alpha (HSP90A) | LAP-2 | LPS-associated protein 2 | Lipopolysaccharide-associated protein 2 | Renal carcinoma antigen NY-REN-38 | heat shock protein 90kDa alpha (cytosolic), class A member 1 isoform 2 |
|---|
| Type: | Molecular Chaperone |
|---|
| Mol. Mass.: | 84623.45 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P07900 |
|---|
| Residue: | 732 |
|---|
| Sequence: | MPEETQTQDQPMEEEEVETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIR
YESLTDPSKLDSGKELHINLIPNKQDRTLTIVDTGIGMTKADLINNLGTIAKSGTKAFME
ALQAGADISMIGQFGVGFYSAYLVAEKVTVITKHNDDEQYAWESSAGGSFTVRTDTGEPM
GRGTKVILHLKEDQTEYLEERRIKEIVKKHSQFIGYPITLFVEKERDKEVSDDEAEEKED
KEEEKEKEEKESEDKPEIEDVGSDEEEEKKDGDKKKKKKIKEKYIDQEELNKTKPIWTRN
PDDITNEEYGEFYKSLTNDWEDHLAVKHFSVEGQLEFRALLFVPRRAPFDLFENRKKKNN
IKLYVRRVFIMDNCEELIPEYLNFIRGVVDSEDLPLNISREMLQQSKILKVIRKNLVKKC
LELFTELAEDKENYKKFYEQFSKNIKLGIHEDSQNRKKLSELLRYYTSASGDEMVSLKDY
CTRMKENQKHIYYITGETKDQVANSAFVERLRKHGLEVIYMIEPIDEYCVQQLKEFEGKT
LVSVTKEGLELPEDEEEKKKQEEKKTKFENLCKIMKDILEKKVEKVVVSNRLVTSPCCIV
TSTYGWTANMERIMKAQALRDNSTMGYMAAKKHLEINPDHSIIETLRQKAEADKNDKSVK
DLVILLYETALLSSGFSLEDPQTHANRIYRMIKLGLGIDEDDPTADDTSAAVTEEMPPLE
GDDDTSRMEEVD
|
|
|
|---|
| BDBM50210933 |
|---|
| n/a |
|---|
| Name | BDBM50210933 |
|---|
| Synonyms: | CHEMBL3935883 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C17H19NO3S |
|---|
| Mol. Mass. | 317.403 |
|---|
| SMILES | CC(C)c1cc(C(=O)N2CCc3sccc3C2)c(O)cc1O |
|---|
| Structure | 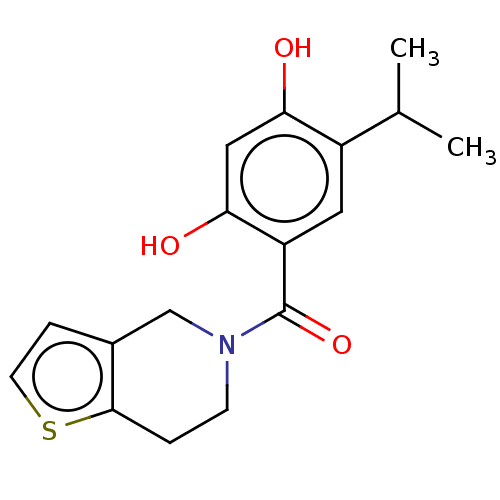
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Jeong, JH; Oh, YJ; Lho, Y; Park, SY; Liu, KH; Ha, E; Seo, YH Targeting the entry region of Hsp90's ATP binding pocket with a novel 6,7-dihydrothieno[3,2-c]pyridin-5(4H)-yl amide. Eur J Med Chem124:1069-1080 (2016) [PubMed] Article
Jeong, JH; Oh, YJ; Lho, Y; Park, SY; Liu, KH; Ha, E; Seo, YH Targeting the entry region of Hsp90's ATP binding pocket with a novel 6,7-dihydrothieno[3,2-c]pyridin-5(4H)-yl amide. Eur J Med Chem124:1069-1080 (2016) [PubMed] Article