

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Induced myeloid leukemia cell differentiation protein Mcl-1 | ||
| Ligand | BDBM286515 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | Bak Peptide Binding Assay | ||
| pH | 7.5±n/a | ||
| Temperature | 298.15±n/a K | ||
| IC50 | <100±n/a nM | ||
| Comments | extracted | ||
| Citation |  Chen, CH; Chen, Z; Fortanet, JG; Grunenfelder, D; Karki, R; Kato, M; LaMarche, MJ; Perez, LB; Stams, TM; Williams, S 1-pyridazin-/triazin-3-yl-piper(-azine)/idine/pyrolidine derivatives and compositions thereof for inhibiting the activity of SHP2 US Patent US10093646 Publication Date 10/9/2018 Chen, CH; Chen, Z; Fortanet, JG; Grunenfelder, D; Karki, R; Kato, M; LaMarche, MJ; Perez, LB; Stams, TM; Williams, S 1-pyridazin-/triazin-3-yl-piper(-azine)/idine/pyrolidine derivatives and compositions thereof for inhibiting the activity of SHP2 US Patent US10093646 Publication Date 10/9/2018 | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Induced myeloid leukemia cell differentiation protein Mcl-1 | |||
| Name: | Induced myeloid leukemia cell differentiation protein Mcl-1 | ||
| Synonyms: | BCL2L3 | Bcl-2-like protein 3 | Bcl-2-like protein 3 (Mcl-1) | Bcl-2-related protein EAT/mcl1 | Bcl2-L-3 | Induced myeloid leukemia cell differentiation protein (Mcl-1) | MCL1 | MCL1_HUMAN | Mcl-1 | Myeloid Cell factor-1 (Mcl-1) | Myeloid cell leukemia sequence 1 (BCL2-related) | mcl1/EAT | ||
| Type: | Membrane; Single-pass membrane protein | ||
| Mol. Mass.: | 37332.87 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q07820 | ||
| Residue: | 350 | ||
| Sequence: |
| ||
| BDBM286515 | |||
| n/a | |||
| Name | BDBM286515 | ||
| Synonyms: | 3-(3-(4-chloro-3,5-dimethylphenoxy)propyl)-7-(3-(hydroxymethyl)-1,5-dimethyl-1H-pyrazol-4-yl)-N-(m-tolylsulfonyl)-1H-indole-2-carboxamide | US10093640, Example 204 | US10844032, Example 204 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C33H35ClN4O5S | ||
| Mol. Mass. | 635.173 | ||
| SMILES | Cc1c(c(CO)nn1C)-c1cccc2c(CCCOc3cc(C)c(Cl)c(C)c3)c([nH]c12)C(=O)NS(=O)(=O)c1cccc(C)c1 |(-1.46,-5.28,;-2.83,-6.07,;-4.12,-5.14,;-5.4,-6.07,;-6.77,-5.28,;-8.15,-6.07,;-4.91,-7.58,;-3.32,-7.58,;-2.53,-8.95,;-4.12,-3.55,;-5.45,-2.78,;-5.45,-1.24,;-4.12,-.47,;-2.78,-1.24,;-1.32,-.77,;-.92,.72,;.57,1.12,;.97,2.6,;2.45,3,;2.85,4.49,;1.76,5.58,;2.16,7.07,;1.07,8.16,;3.65,7.47,;4.05,8.95,;4.74,6.38,;6.22,6.77,;4.34,4.89,;-.41,-2.01,;-1.32,-3.26,;-2.78,-2.78,;1.13,-2.01,;1.9,-3.35,;1.9,-.68,;3.44,-.68,;3.44,-2.22,;3.44,.86,;4.98,-.68,;5.77,.69,;7.35,.69,;8.15,-.68,;7.35,-2.05,;8.15,-3.43,;5.77,-2.05,)| | ||
| Structure | 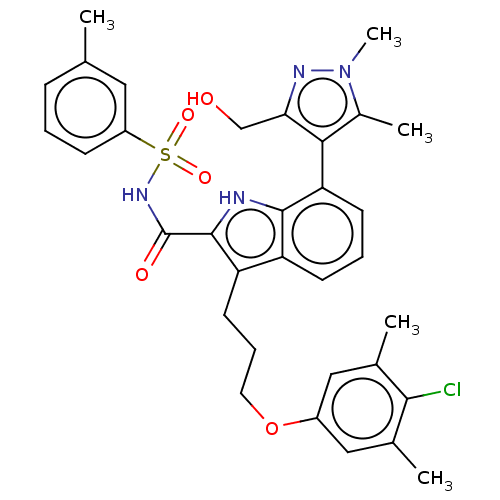
| ||