

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Diacylglycerol O-acyltransferase 1 | ||
| Ligand | BDBM50502586 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1808007 (CHEMBL4307366) | ||
| EC50 | 71±n/a nM | ||
| Citation |  Harrison, TJ; Bauer, D; Berdichevsky, A; Chen, X; Duvadie, R; Hoogheem, B; Hatsis, P; Liu, Q; Mao, J; Miduturu, V; Rocheford, E; Zecri, F; Zessis, R; Zheng, R; Zhu, Q; Streeper, R; Patel, SJ Successful Strategies for Mitigation of a Preclinical Signal for Phototoxicity in a DGAT1 Inhibitor. ACS Med Chem Lett10:1128-1133 (2019) [PubMed] Article Harrison, TJ; Bauer, D; Berdichevsky, A; Chen, X; Duvadie, R; Hoogheem, B; Hatsis, P; Liu, Q; Mao, J; Miduturu, V; Rocheford, E; Zecri, F; Zessis, R; Zheng, R; Zhu, Q; Streeper, R; Patel, SJ Successful Strategies for Mitigation of a Preclinical Signal for Phototoxicity in a DGAT1 Inhibitor. ACS Med Chem Lett10:1128-1133 (2019) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Diacylglycerol O-acyltransferase 1 | |||
| Name: | Diacylglycerol O-acyltransferase 1 | ||
| Synonyms: | DGAT1_MOUSE | Dgat | Dgat1 | Diacyl Glycerolacyltransferase 1 (DGAT-1) | Diacylglycerol O-acyltransferase 1 (DGAT1) | Diglyceride acyltransferase | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 56810.61 | ||
| Organism: | Mus musculus (mouse) | ||
| Description: | In this assay, recombinant mouse DGAT-1 containing an N-terminal His6-epitope tag was produced in the baculovirus expression system. | ||
| Residue: | 498 | ||
| Sequence: |
| ||
| BDBM50502586 | |||
| n/a | |||
| Name | BDBM50502586 | ||
| Synonyms: | LCQ-908-NXA | LCQ-908NXA | LCQ908-NXA | Pradigastat | ||
| Type | Small organic molecule | ||
| Emp. Form. | C25H24F3N3O2 | ||
| Mol. Mass. | 455.4722 | ||
| SMILES | OC(=O)C[C@H]1CC[C@@H](CC1)c1ccc(cc1)-c1ccc(Nc2ccc(nc2)C(F)(F)F)cn1 |r,wU:4.3,wD:7.10,(-2.34,-12.23,;-2.34,-10.69,;-3.68,-9.92,;-1.01,-9.92,;-1.01,-8.38,;-2.34,-7.61,;-2.34,-6.07,;-1.01,-5.3,;.32,-6.07,;.32,-7.61,;-1.01,-3.76,;-2.34,-2.99,;-2.34,-1.45,;-1.01,-.68,;.32,-1.45,;.32,-2.99,;-1.01,.86,;-2.34,1.63,;-2.34,3.17,;-1.01,3.94,;-1.01,5.48,;.32,6.25,;.32,7.79,;1.66,8.56,;2.99,7.79,;2.99,6.25,;1.66,5.48,;4.32,8.56,;5.66,9.33,;5.09,7.23,;3.55,9.9,;.32,3.17,;.32,1.63,)| | ||
| Structure | 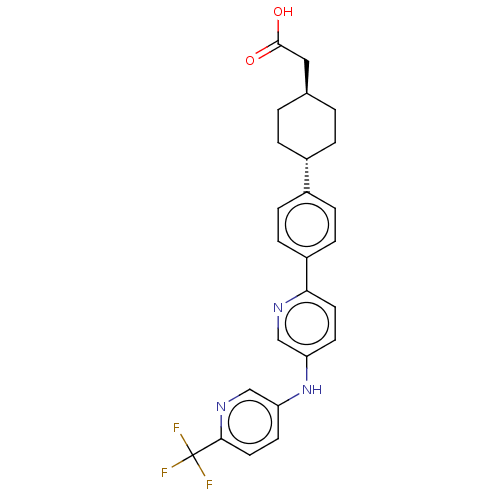
| ||