| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Cathepsin D |
|---|
| Ligand | BDBM50012632 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1980875 (CHEMBL4614137) |
|---|
| IC50 | 15700±n/a nM |
|---|
| Citation |  Winneroski, LL; Erickson, JA; Green, SJ; Lopez, JE; Stout, SL; Porter, WJ; Timm, DE; Audia, JE; Barberis, M; Beck, JP; Boggs, LN; Borders, AR; Boyer, RD; Brier, RA; Hembre, EJ; Hendle, J; Garcia-Losada, P; Minguez, JM; Mathes, BM; May, PC; Monk, SA; Rankovic, Z; Shi, Y; Watson, BM; Yang, Z; Mergott, DJ Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints. Bioorg Med Chem28:0 (2020) [PubMed] Article Winneroski, LL; Erickson, JA; Green, SJ; Lopez, JE; Stout, SL; Porter, WJ; Timm, DE; Audia, JE; Barberis, M; Beck, JP; Boggs, LN; Borders, AR; Boyer, RD; Brier, RA; Hembre, EJ; Hendle, J; Garcia-Losada, P; Minguez, JM; Mathes, BM; May, PC; Monk, SA; Rankovic, Z; Shi, Y; Watson, BM; Yang, Z; Mergott, DJ Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints. Bioorg Med Chem28:0 (2020) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Cathepsin D |
|---|
| Name: | Cathepsin D |
|---|
| Synonyms: | CATD_HUMAN | CPSD | CTSD | Cathepsin D [Precursor] | Cathepsin D heavy chain | Cathepsin D light chain | Cathepsin D precursor |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 44551.72 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Human proCathepsin D (SwissProt accession number P07339) was expressed in Sf9 cells, purified, and autoactivated. |
|---|
| Residue: | 412 |
|---|
| Sequence: | MQPSSLLPLALCLLAAPASALVRIPLHKFTSIRRTMSEVGGSVEDLIAKGPVSKYSQAVP
AVTEGPIPEVLKNYMDAQYYGEIGIGTPPQCFTVVFDTGSSNLWVPSIHCKLLDIACWIH
HKYNSDKSSTYVKNGTSFDIHYGSGSLSGYLSQDTVSVPCQSASSASALGGVKVERQVFG
EATKQPGITFIAAKFDGILGMAYPRISVNNVLPVFDNLMQQKLVDQNIFSFYLSRDPDAQ
PGGELMLGGTDSKYYKGSLSYLNVTRKAYWQVHLDQVEVASGLTLCKEGCEAIVDTGTSL
MVGPVDEVRELQKAIGAVPLIQGEYMIPCEKVSTLPAITLKLGGKGYKLSPEDYTLKVSQ
AGKTLCLSGFMGMDIPPPSGPLWILGDVFIGRYYTVFDRDNNRVGFAEAARL
|
|
|
|---|
| BDBM50012632 |
|---|
| n/a |
|---|
| Name | BDBM50012632 |
|---|
| Synonyms: | CHEMBL2333941 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C15H14F2N4S |
|---|
| Mol. Mass. | 320.36 |
|---|
| SMILES | C[C@]1(CCSC(N)=N1)c1cc(c(F)cc1F)-c1cncnc1 |r,c:6| |
|---|
| Structure | 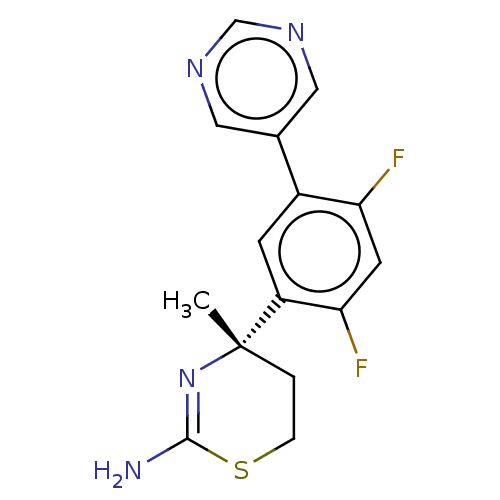
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Winneroski, LL; Erickson, JA; Green, SJ; Lopez, JE; Stout, SL; Porter, WJ; Timm, DE; Audia, JE; Barberis, M; Beck, JP; Boggs, LN; Borders, AR; Boyer, RD; Brier, RA; Hembre, EJ; Hendle, J; Garcia-Losada, P; Minguez, JM; Mathes, BM; May, PC; Monk, SA; Rankovic, Z; Shi, Y; Watson, BM; Yang, Z; Mergott, DJ Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints. Bioorg Med Chem28:0 (2020) [PubMed] Article
Winneroski, LL; Erickson, JA; Green, SJ; Lopez, JE; Stout, SL; Porter, WJ; Timm, DE; Audia, JE; Barberis, M; Beck, JP; Boggs, LN; Borders, AR; Boyer, RD; Brier, RA; Hembre, EJ; Hendle, J; Garcia-Losada, P; Minguez, JM; Mathes, BM; May, PC; Monk, SA; Rankovic, Z; Shi, Y; Watson, BM; Yang, Z; Mergott, DJ Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints. Bioorg Med Chem28:0 (2020) [PubMed] Article