| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Interleukin-1 receptor-associated kinase 4 |
|---|
| Ligand | BDBM50262366 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1991895 (CHEMBL4625630) |
|---|
| IC50 | 11±n/a nM |
|---|
| Citation |  Nair, S; Kumar, SR; Paidi, VR; Sistla, R; Kantheti, D; Polimera, SR; Thangavel, S; Mukherjee, AJ; Das, M; Bhide, RS; Pitts, WJ; Murugesan, N; Dudhgoankar, S; Nagar, J; Subramani, S; Mazumder, D; Carman, JA; Holloway, DA; Li, X; Fereshteh, MP; Ruepp, S; Palanisamy, K; Mariappan, TT; Maddi, S; Saxena, A; Elzinga, P; Chimalakonda, A; Ruan, Q; Ghosh, K; Bose, S; Sack, J; Yan, C; Kiefer, SE; Xie, D; Newitt, JA; Saravanakumar, SP; Rampulla, RA; Barrish, JC; Carter, PH; Hynes, J Optimization of Nicotinamides as Potent and Selective IRAK4 Inhibitors with Efficacy in a Murine Model of Psoriasis. ACS Med Chem Lett11:1402-1409 (2020) [PubMed] Article Nair, S; Kumar, SR; Paidi, VR; Sistla, R; Kantheti, D; Polimera, SR; Thangavel, S; Mukherjee, AJ; Das, M; Bhide, RS; Pitts, WJ; Murugesan, N; Dudhgoankar, S; Nagar, J; Subramani, S; Mazumder, D; Carman, JA; Holloway, DA; Li, X; Fereshteh, MP; Ruepp, S; Palanisamy, K; Mariappan, TT; Maddi, S; Saxena, A; Elzinga, P; Chimalakonda, A; Ruan, Q; Ghosh, K; Bose, S; Sack, J; Yan, C; Kiefer, SE; Xie, D; Newitt, JA; Saravanakumar, SP; Rampulla, RA; Barrish, JC; Carter, PH; Hynes, J Optimization of Nicotinamides as Potent and Selective IRAK4 Inhibitors with Efficacy in a Murine Model of Psoriasis. ACS Med Chem Lett11:1402-1409 (2020) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Interleukin-1 receptor-associated kinase 4 |
|---|
| Name: | Interleukin-1 receptor-associated kinase 4 |
|---|
| Synonyms: | IRAK-4 | IRAK4 | IRAK4_HUMAN | Interleukin-1 receptor-associated kinase 4 (IRAK-4) | Interleukin-1 receptor-associated kinase 4 (IRAK4) | Renal carcinoma antigen NY-REN-64 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 51519.08 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9NWZ3 |
|---|
| Residue: | 460 |
|---|
| Sequence: | MNKPITPSTYVRCLNVGLIRKLSDFIDPQEGWKKLAVAIKKPSGDDRYNQFHIRRFEALL
QTGKSPTSELLFDWGTTNCTVGDLVDLLIQNEFFAPASLLLPDAVPKTANTLPSKEAITV
QQKQMPFCDKDRTLMTPVQNLEQSYMPPDSSSPENKSLEVSDTRFHSFSFYELKNVTNNF
DERPISVGGNKMGEGGFGVVYKGYVNNTTVAVKKLAAMVDITTEELKQQFDQEIKVMAKC
QHENLVELLGFSSDGDDLCLVYVYMPNGSLLDRLSCLDGTPPLSWHMRCKIAQGAANGIN
FLHENHHIHRDIKSANILLDEAFTAKISDFGLARASEKFAQTVMTSRIVGTTAYMAPEAL
RGEITPKSDIYSFGVVLLEIITGLPAVDEHREPQLLLDIKEEIEDEEKTIEDYIDKKMND
ADSTSVEAMYSVASQCLHEKKNKRPDIKKVQQLLQEMTAS
|
|
|
|---|
| BDBM50262366 |
|---|
| n/a |
|---|
| Name | BDBM50262366 |
|---|
| Synonyms: | CHEMBL4079743 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C23H23N5O2S |
|---|
| Mol. Mass. | 433.526 |
|---|
| SMILES | CNC(=O)c1cnc(Nc2ccc3ncsc3c2)cc1N[C@H](CO)Cc1ccccc1 |r| |
|---|
| Structure | 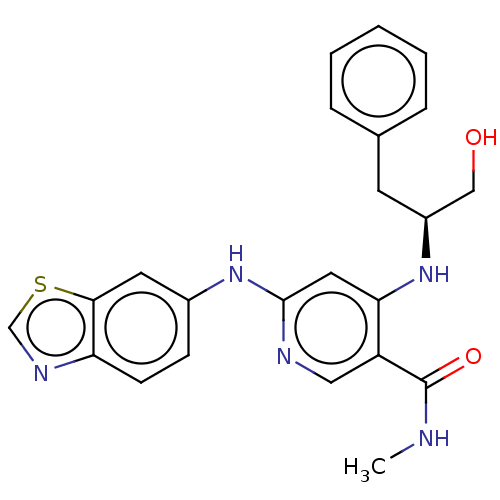
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Nair, S; Kumar, SR; Paidi, VR; Sistla, R; Kantheti, D; Polimera, SR; Thangavel, S; Mukherjee, AJ; Das, M; Bhide, RS; Pitts, WJ; Murugesan, N; Dudhgoankar, S; Nagar, J; Subramani, S; Mazumder, D; Carman, JA; Holloway, DA; Li, X; Fereshteh, MP; Ruepp, S; Palanisamy, K; Mariappan, TT; Maddi, S; Saxena, A; Elzinga, P; Chimalakonda, A; Ruan, Q; Ghosh, K; Bose, S; Sack, J; Yan, C; Kiefer, SE; Xie, D; Newitt, JA; Saravanakumar, SP; Rampulla, RA; Barrish, JC; Carter, PH; Hynes, J Optimization of Nicotinamides as Potent and Selective IRAK4 Inhibitors with Efficacy in a Murine Model of Psoriasis. ACS Med Chem Lett11:1402-1409 (2020) [PubMed] Article
Nair, S; Kumar, SR; Paidi, VR; Sistla, R; Kantheti, D; Polimera, SR; Thangavel, S; Mukherjee, AJ; Das, M; Bhide, RS; Pitts, WJ; Murugesan, N; Dudhgoankar, S; Nagar, J; Subramani, S; Mazumder, D; Carman, JA; Holloway, DA; Li, X; Fereshteh, MP; Ruepp, S; Palanisamy, K; Mariappan, TT; Maddi, S; Saxena, A; Elzinga, P; Chimalakonda, A; Ruan, Q; Ghosh, K; Bose, S; Sack, J; Yan, C; Kiefer, SE; Xie, D; Newitt, JA; Saravanakumar, SP; Rampulla, RA; Barrish, JC; Carter, PH; Hynes, J Optimization of Nicotinamides as Potent and Selective IRAK4 Inhibitors with Efficacy in a Murine Model of Psoriasis. ACS Med Chem Lett11:1402-1409 (2020) [PubMed] Article