| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Peroxisome proliferator-activated receptor alpha |
|---|
| Ligand | BDBM50584495 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2160008 (CHEMBL5044758) |
|---|
| EC50 | 382±n/a nM |
|---|
| Citation |  Feng, Z; Xiang, J; Liu, H; Li, J; Xu, X; Sun, G; Zheng, R; Zhang, S; Liu, J; Yang, S; Xu, Q; Wen, X; Yuan, H; Sun, H; Dai, L Design, Synthesis, and Biological Evaluation of Triazolone Derivatives as Potent PPAR?/? Dual Agonists for the Treatment of Nonalcoholic Steatohepatitis. J Med Chem65:2571-2592 (2022) [PubMed] Article Feng, Z; Xiang, J; Liu, H; Li, J; Xu, X; Sun, G; Zheng, R; Zhang, S; Liu, J; Yang, S; Xu, Q; Wen, X; Yuan, H; Sun, H; Dai, L Design, Synthesis, and Biological Evaluation of Triazolone Derivatives as Potent PPAR?/? Dual Agonists for the Treatment of Nonalcoholic Steatohepatitis. J Med Chem65:2571-2592 (2022) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Peroxisome proliferator-activated receptor alpha |
|---|
| Name: | Peroxisome proliferator-activated receptor alpha |
|---|
| Synonyms: | NR1C1 | Nuclear receptor subfamily 1 group C member 1 | PPAR | PPAR alpha/gamma | PPAR-alpha | PPARA | PPARA_HUMAN | Peroxisome Proliferator-Activated Receptor alpha | Peroxisome proliferator-activated receptor | Peroxisome proliferator-activated receptor alpha (PPAR alpha) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 52222.08 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q07869 |
|---|
| Residue: | 468 |
|---|
| Sequence: | MVDTESPLCPLSPLEAGDLESPLSEEFLQEMGNIQEISQSIGEDSSGSFGFTEYQYLGSC
PGSDGSVITDTLSPASSPSSVTYPVVPGSVDESPSGALNIECRICGDKASGYHYGVHACE
GCKGFFRRTIRLKLVYDKCDRSCKIQKKNRNKCQYCRFHKCLSVGMSHNAIRFGRMPRSE
KAKLKAEILTCEHDIEDSETADLKSLAKRIYEAYLKNFNMNKVKARVILSGKASNNPPFV
IHDMETLCMAEKTLVAKLVANGIQNKEAEVRIFHCCQCTSVETVTELTEFAKAIPGFANL
DLNDQVTLLKYGVYEAIFAMLSSVMNKDGMLVAYGNGFITREFLKSLRKPFCDIMEPKFD
FAMKFNALELDDSDISLFVAAIICCGDRPGLLNVGHIEKMQEGIVHVLRLHLQSNHPDDI
FLFPKLLQKMADLRQLVTEHAQLVQIIKKTESDAALHPLLQEIYRDMY
|
|
|
|---|
| BDBM50584495 |
|---|
| n/a |
|---|
| Name | BDBM50584495 |
|---|
| Synonyms: | CHEMBL5077842 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C21H17F6N3O5 |
|---|
| Mol. Mass. | 505.3672 |
|---|
| SMILES | CC(C)(Oc1ccc(Cn2ncn(-c3ccc(cc3)C(F)(F)F)c2=O)cc1OC(F)(F)F)C(O)=O |
|---|
| Structure | 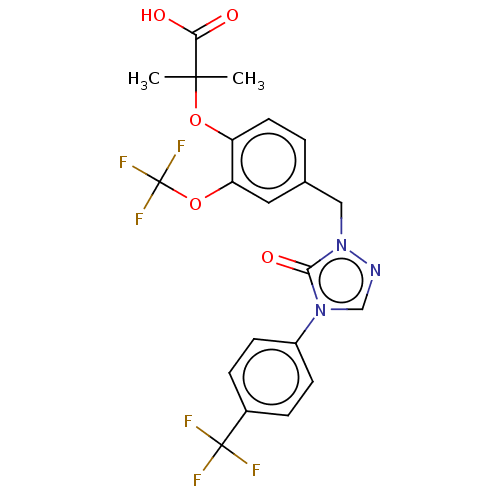
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Feng, Z; Xiang, J; Liu, H; Li, J; Xu, X; Sun, G; Zheng, R; Zhang, S; Liu, J; Yang, S; Xu, Q; Wen, X; Yuan, H; Sun, H; Dai, L Design, Synthesis, and Biological Evaluation of Triazolone Derivatives as Potent PPAR?/? Dual Agonists for the Treatment of Nonalcoholic Steatohepatitis. J Med Chem65:2571-2592 (2022) [PubMed] Article
Feng, Z; Xiang, J; Liu, H; Li, J; Xu, X; Sun, G; Zheng, R; Zhang, S; Liu, J; Yang, S; Xu, Q; Wen, X; Yuan, H; Sun, H; Dai, L Design, Synthesis, and Biological Evaluation of Triazolone Derivatives as Potent PPAR?/? Dual Agonists for the Treatment of Nonalcoholic Steatohepatitis. J Med Chem65:2571-2592 (2022) [PubMed] Article